| (28) B-CELL | (8) BREAST | (17) CELL-LINE | (3) CERVIX | (1) HELA | (6) LIVER | (1) OTHER | (8) SKIN | (4) UTERUS |
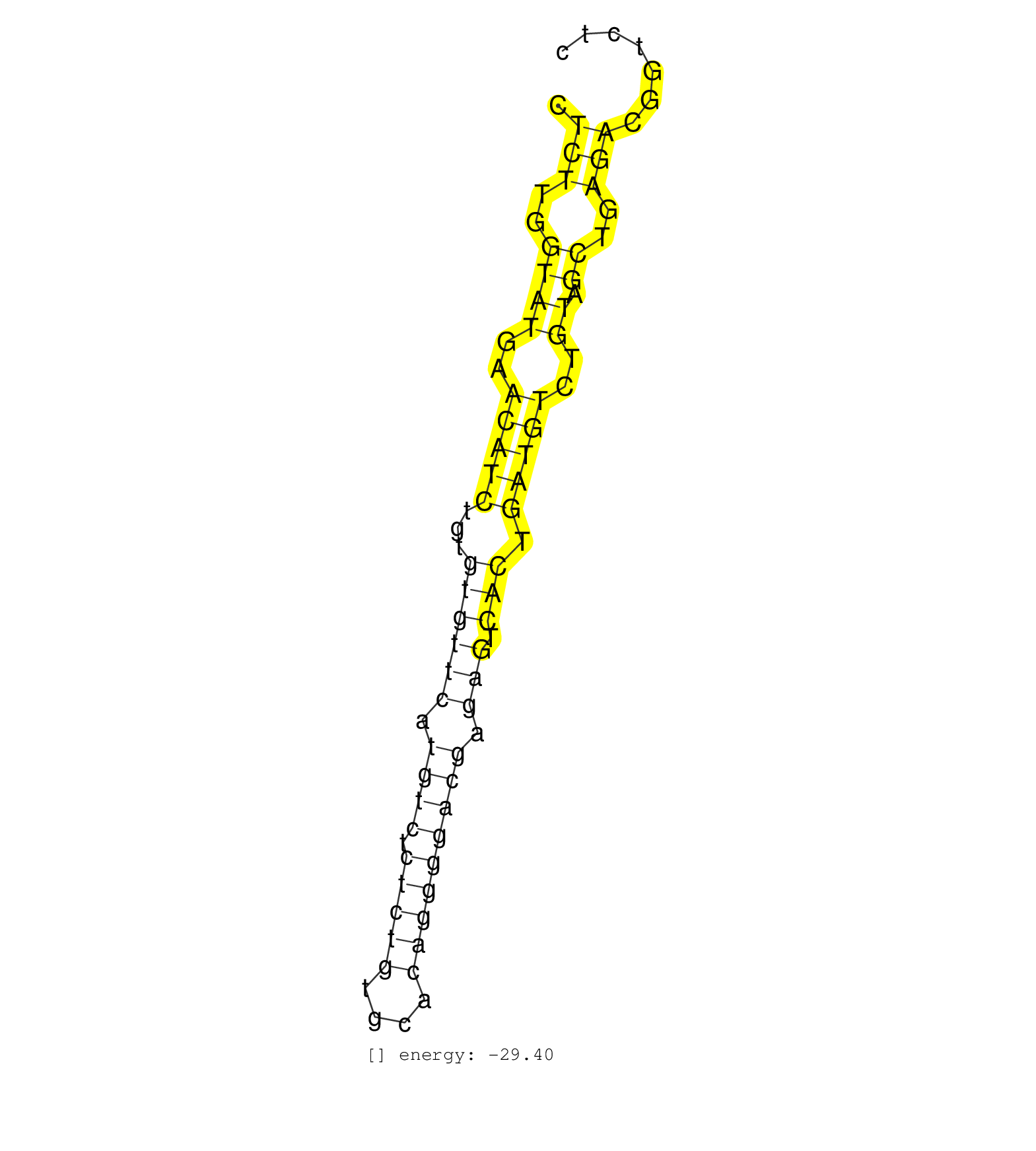
| CTGGCCCGGGAGTCTGGAGACCTTGGTTCTAGTCTTGGGTCTGCCACCAGCTCTTGGTATGAACATCTGTGTGTTCATGTCTCTCTGTGCACAGGGGACGAGAGTCACTGATGTCTGTAGCTGAGACGGTCTCTCAAGTGAGGGCCTACTATGTGCCGCTCAGTAACTGGGACTTTA ...................................................(((..((((..(((((...((((((.((((.(((((....))))))))).))).))).)))))..)).))..)))......................................................... ..................................................51................................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR040023(GSM532908) G575T. (cervix) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR038862(GSM458545) MM472. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR040034(GSM532919) G001N. (cervix) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189784 | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGG................................................ | 26 | 1 | 2216.00 | 2216.00 | 769.00 | 230.00 | 241.00 | 90.00 | 132.00 | 79.00 | 90.00 | 98.00 | 51.00 | 75.00 | 51.00 | 65.00 | 20.00 | 35.00 | 41.00 | 24.00 | 14.00 | 4.00 | 14.00 | 6.00 | 14.00 | 4.00 | 6.00 | 4.00 | 4.00 | 4.00 | 2.00 | - | 5.00 | 3.00 | 6.00 | 1.00 | - | 1.00 | 2.00 | 3.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | 2.00 | 1.00 | 2.00 | 2.00 | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGG................................................ | 27 | 1 | 1049.00 | 1049.00 | 364.00 | 121.00 | 68.00 | 84.00 | 47.00 | 80.00 | 47.00 | 17.00 | 53.00 | 9.00 | 14.00 | 26.00 | 4.00 | 21.00 | 27.00 | 26.00 | 2.00 | 10.00 | - | - | - | 4.00 | 2.00 | - | 2.00 | - | 1.00 | - | - | 2.00 | - | - | 2.00 | 2.00 | 1.00 | - | 1.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACG................................................. | 25 | 1 | 545.00 | 545.00 | 147.00 | 44.00 | 50.00 | 18.00 | 23.00 | 35.00 | 30.00 | 33.00 | 28.00 | 25.00 | 17.00 | 14.00 | 31.00 | 9.00 | - | 8.00 | 8.00 | - | 2.00 | 4.00 | 2.00 | 1.00 | - | 2.00 | - | 1.00 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACG................................................. | 26 | 1 | 364.00 | 364.00 | 119.00 | 32.00 | 30.00 | 19.00 | 10.00 | 15.00 | 20.00 | 13.00 | 24.00 | 15.00 | 17.00 | 8.00 | 15.00 | 2.00 | - | 4.00 | 4.00 | 2.00 | - | 6.00 | - | - | - | - | - | - | - | 3.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTGATGTCTGTAGCTGAGACGG................................................ | 23 | 1 | 46.00 | 46.00 | 16.00 | 3.00 | 5.00 | 2.00 | 5.00 | 1.00 | 1.00 | 5.00 | 1.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAagg................................................ | 26 | AGG | 17.00 | 5.00 | 3.00 | - | - | 6.00 | - | 1.00 | 1.00 | - | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGT............................................... | 27 | 1 | 10.00 | 10.00 | 7.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGaa.............................................. | 28 | AA | 9.00 | 2216.00 | 5.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGa................................................ | 26 | A | 7.00 | 545.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGccgg................................................ | 26 | CCGG | 6.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGA................................................... | 23 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGa............................................... | 27 | A | 5.00 | 2216.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGtcgg................................................ | 26 | TCGG | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGccg................................................. | 25 | CCG | 4.00 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAtacg................................................. | 26 | TACG | 4.00 | 0.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGA................................................... | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGGa............................................... | 28 | A | 4.00 | 1049.00 | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGGT............................................... | 28 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACag................................................ | 27 | AG | 4.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACcg................................................ | 26 | CG | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACtg................................................ | 26 | TG | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TCACTGATGTCTGTAGCTGAGACGG................................................ | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGccgg................................................ | 27 | CCGG | 3.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAag................................................. | 25 | AG | 3.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGa................................................ | 27 | A | 3.00 | 364.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTGATGTCTGTAGCTGAGACG................................................. | 22 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAtgg................................................ | 26 | TGG | 3.00 | 5.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAC.................................................. | 24 | 1 | 3.00 | 3.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAtacg................................................. | 25 | TACG | 3.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACag................................................ | 26 | AG | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGgcgg................................................ | 26 | GCGG | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGAtg................................................. | 26 | TG | 2.00 | 4.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGg............................................... | 27 | G | 2.00 | 2216.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CTGATGTCTGTAGCTGAGACGG................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACc................................................. | 25 | C | 2.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGAtgg................................................ | 27 | TGG | 2.00 | 4.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGTCTGTAGCTGAGACGG................................................ | 18 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGt................................................ | 26 | T | 2.00 | 545.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGAagg................................................ | 27 | AGG | 2.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGtcg................................................. | 25 | TCG | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAG.................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGAggg................................................ | 27 | GGG | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAg.................................................. | 24 | G | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGc................................................ | 26 | C | 1.00 | 545.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAaa................................................. | 25 | AA | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGtcgg.................................................. | 25 | TCGG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACt................................................. | 26 | T | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAttg................................................ | 26 | TTG | 1.00 | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAa.................................................. | 24 | A | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGAag................................................. | 26 | AG | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAcgg.................................................. | 25 | CGG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACc................................................. | 26 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTGATGTCTGTAGCTGAGAagg................................................ | 23 | AGG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGc................................................ | 27 | C | 1.00 | 364.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGc............................................... | 27 | C | 1.00 | 2216.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGccg................................................. | 26 | CCG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACcg................................................ | 27 | CG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGtcgg................................................ | 27 | TCGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTtag.................................................... | 23 | TAG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTGATGTCTGTAGCTGAGACGGaa.............................................. | 25 | AA | 1.00 | 46.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACtgt............................................... | 27 | TGT | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GATGTCTGTAGCTGAGACGG................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGgcg................................................. | 25 | GCG | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTtgg.................................................... | 22 | TGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAtg................................................. | 25 | TG | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAac................................................. | 25 | AC | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTtag.................................................... | 22 | TAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GAGTCTGGAGACCTTcga...................................................................................................................................................... | 18 | CGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGCTCTTGGTATGAACATC.............................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAaga................................................ | 26 | AGA | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAtcg................................................ | 26 | TCG | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GGAGTCTGGAGACCTgct....................................................................................................................................................... | 18 | GCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGtcg................................................. | 26 | TCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................GAGTCACTGATGTCTGTAGCTGAGACGG................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGccc................................................. | 26 | CCC | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................GAGTCACTGATGTCTGTAGCTGAGACG................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGtcgt................................................ | 26 | TCGT | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGcagg................................................ | 26 | CAGG | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CACTGATGTCTGTAGCTGAGACGG................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGAggg................................................ | 26 | GGG | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGaa............................................... | 28 | AA | 1.00 | 364.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTCACTGATGTCTGTAGCTGAGACGGTCTCTa.......................................... | 32 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GGCCCGGGAGTCTGGggg............................................................................................................................................................. | 18 | GGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGgcg................................................. | 26 | GCG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTCACTGATGTCTGTAGCTGAGACGGTa.............................................. | 29 | A | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGGCCCGGGAGTCTGGAGACCTTGGTTCTAGTCTTGGGTCTGCCACCAGCTCTTGGTATGAACATCTGTGTGTTCATGTCTCTCTGTGCACAGGGGACGAGAGTCACTGATGTCTGTAGCTGAGACGGTCTCTCAAGTGAGGGCCTACTATGTGCCGCTCAGTAACTGGGACTTTA ...................................................(((..((((..(((((...((((((.((((.(((((....))))))))).))).))).)))))..)).))..)))......................................................... ..................................................51................................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR040023(GSM532908) G575T. (cervix) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR038862(GSM458545) MM472. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR040034(GSM532919) G001N. (cervix) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189784 | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR038859(GSM458542) MM386. (cell line) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................gaccCGGTCTCTCAAGTGA.................................... | 19 | gacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................cagTTGGTATGAACATCTGTGTGTTCATG.................................................................................................. | 29 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................CTCTTGGTATGAACATCTGTGTGTTCATG.................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |