| (15) B-CELL | (2) BRAIN | (1) BREAST | (1) CELL-LINE | (3) HEART | (1) LIVER | (4) OTHER | (1) PLACENTA | (3) SKIN | (3) SPLEEN | (2) THYMUS |
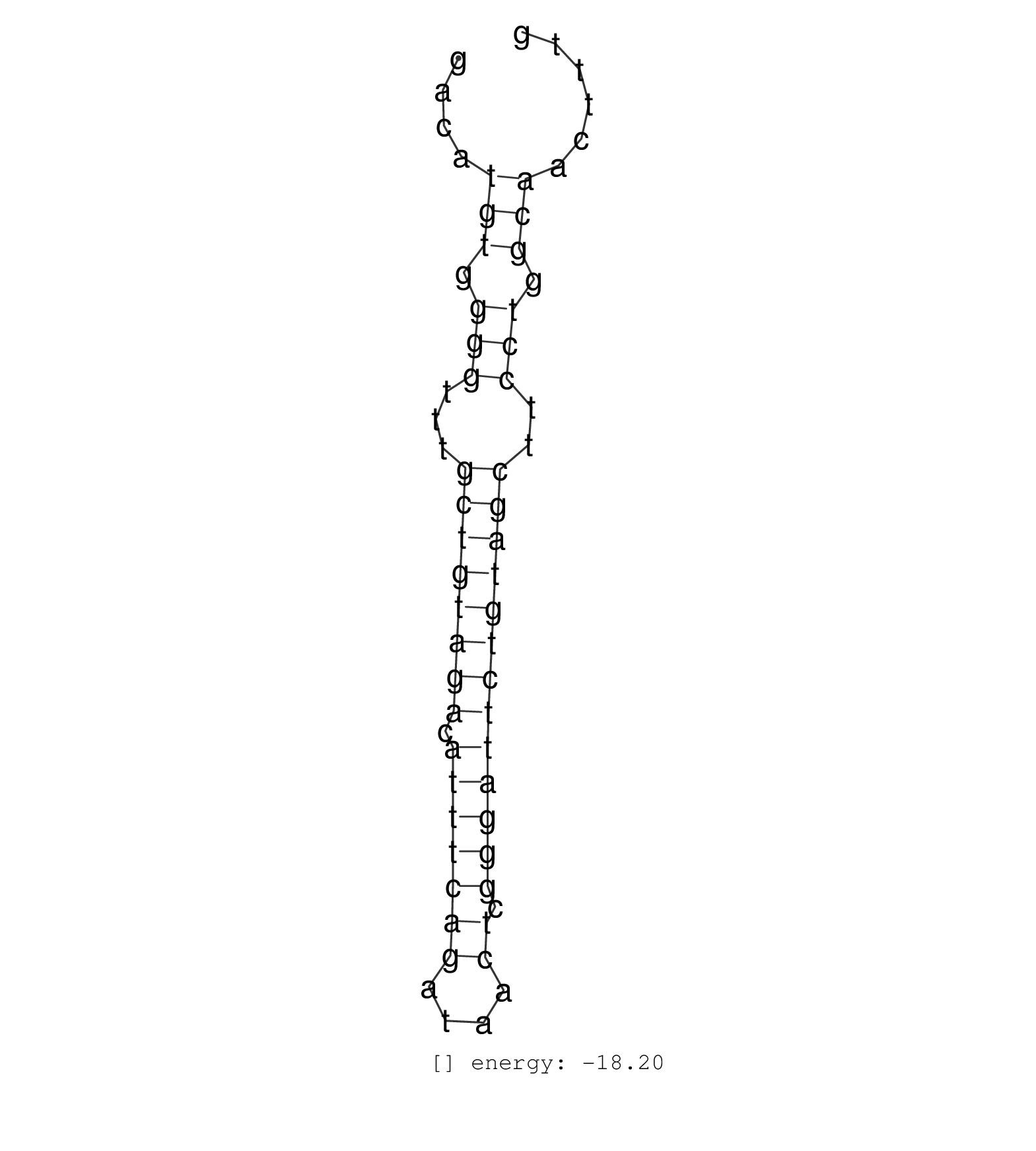
| CAGGAATGTGTGCAAGCTCTTGACTTGTCAAGGCCTTTGCTCACCAGAGTGACATGTGGGGTTTGCTGTAGACATTTCAGATAACTCGGGATTCTGTAGCTTCCTGGCAACTTTGCCCTGACAGCCCCCACTTTGTGGTCCCCTGTGATTTGCCCTCTTAGCTCC ......................................................(((.(((...((((((((.(((((((....)).)))))))))))))..))).)))........................................................ ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139206(SRX050644) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR189785 | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR139219(SRX050655) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR139169(SRX050641) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189784 | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR139198(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139162(SRX049422) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR139215(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR343336 | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR139166(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | TAX577741(Rovira) total RNA. (breast) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR343337 | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........GTGCAAGCTCTTGACTgaag........................................................................................................................................ | 20 | GAAG | 14.00 | 0.00 | - | - | 2.00 | 2.00 | 1.00 | - | - | 3.00 | - | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........TGCAAGCTCTTGACTgaag........................................................................................................................................ | 19 | GAAG | 7.00 | 0.00 | - | - | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GTGCAAGCTCTTGACTgaa......................................................................................................................................... | 19 | GAA | 6.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TGTGCAAGCTCTTGAttga.......................................................................................................................................... | 19 | TTGA | 6.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................GGGATTCTGTAGCTTCCTc........................................................... | 19 | C | 6.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GTGCAAGCTCTTGACTga.......................................................................................................................................... | 18 | GA | 4.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................GATTCTGTAGCTTCCTc........................................................... | 17 | C | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTgctc........................................................... | 19 | GCTC | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTtc............................................................. | 17 | TC | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGTGGGGTTTGCTGTAGACAct......................................................................................... | 22 | CT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGATTCTGTAGCTTCCTc........................................................... | 18 | C | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCgtc........................................................... | 19 | GTC | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTggtc........................................................... | 19 | GGTC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGATTCTGTAGCTTCCTct.......................................................... | 21 | CT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGATTCTGTAGCTTCatc........................................................... | 18 | ATC | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GTGCAAGCTCTTGACTTaagt....................................................................................................................................... | 21 | AAGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GTGTGCAAGCTCTTGACTgaag........................................................................................................................................ | 22 | GAAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCtct........................................................... | 19 | TCT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGATTCTGTAGCTTac............................................................. | 18 | AC | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GAATGTGTGCAAGCTCTTG............................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCgctc.......................................................... | 20 | GCTC | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCCTcgg......................................................... | 21 | CGG | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCCga........................................................... | 19 | GA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTtctc........................................................... | 19 | TCTC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TTGACTTGTCAAGGCCTTTGCTC........................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................GGGATTCTGTAGCTTgc............................................................. | 17 | GC | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGATTCTGTAGCTTCt............................................................. | 18 | T | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GATTCTGTAGCTTCCTca.......................................................... | 18 | CA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCtc............................................................ | 18 | TC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGATTCTGTAGCT................................................................ | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCtcg........................................................... | 19 | TCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGATTCTGTAGCTTCCTca.......................................................... | 21 | CA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCAAGCTCTTGACTgaa......................................................................................................................................... | 18 | GAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCCTcag......................................................... | 21 | CAG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GATTCTGTAGCTTCCTcg.......................................................... | 18 | CG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCCTca.......................................................... | 20 | CA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGATTCTGTAGCTTCCTcagt........................................................ | 21 | CAGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GGGATTCTGTAGCTTCttca.......................................................... | 20 | TTCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................GATAACTCGGGATTCTGTAGCTTCCTG........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGATTCTGTAGCTTCCTGa.......................................................... | 19 | A | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGATTCTGTAGCTTCttct.......................................................... | 19 | TTCT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GTGTGCAAGCTCTTGA.............................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGGAATGTGTGCAAGCTCTTGACTTGTCAAGGCCTTTGCTCACCAGAGTGACATGTGGGGTTTGCTGTAGACATTTCAGATAACTCGGGATTCTGTAGCTTCCTGGCAACTTTGCCCTGACAGCCCCCACTTTGTGGTCCCCTGTGATTTGCCCTCTTAGCTCC ......................................................(((.(((...((((((((.(((((((....)).)))))))))))))..))).)))........................................................ ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139206(SRX050644) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR189785 | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR139219(SRX050655) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR139169(SRX050641) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189784 | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR139198(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139162(SRX049422) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR139215(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR343336 | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR139166(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | TAX577741(Rovira) total RNA. (breast) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR343337 | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................ggGATTTGCCCTCTTAGC... | 18 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................cactCCCTGACAGCCCCCA................................... | 19 | cact | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |