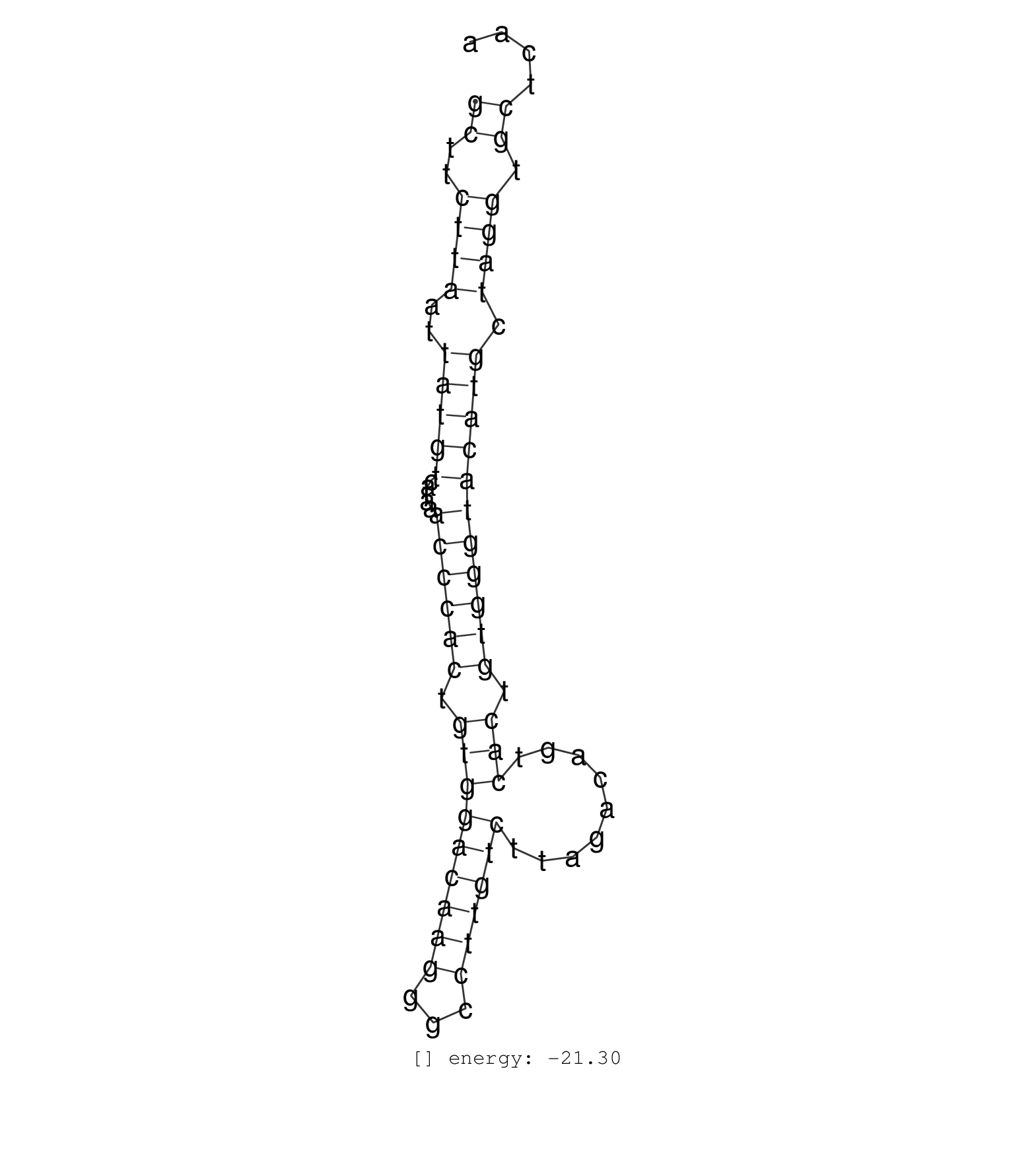
| (1) CELL-LINE | (2) OTHER |
| TACCTGTACCATCTGTTATTACCTCTTAATTCCTTCATGAGTTTGTAGGAGCTTCTTAATTATGTCATAAACCCACTGTGGACAAGGGCCTTGTCTTAGACAGTCACTGTGGGTACATGCTAGGTGCTCAAAAACTTGTTTCAATGATTGATTTCTATATTCATGAGGATCAAAATAATCC ..................................................((..((((..(((((.....((((((.(((((((((...)))))).........))).))))))))))).)))).))...................................................... ..................................................51..............................................................................131................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | SRR189785 | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................ATTTCTATATTCATGccga............ | 19 | CCGA | 1.00 | 0.00 | - | 1.00 | - |
| TACCTGTACCATCTGTTATTACCTCTTAATTCCTTCATGAGTTTGTAGGAGCTTCTTAATTATGTCATAAACCCACTGTGGACAAGGGCCTTGTCTTAGACAGTCACTGTGGGTACATGCTAGGTGCTCAAAAACTTGTTTCAATGATTGATTTCTATATTCATGAGGATCAAAATAATCC ..................................................((..((((..(((((.....((((((.(((((((((...)))))).........))).))))))))))).)))).))...................................................... ..................................................51..............................................................................131................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | SRR189785 | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................taaCATGAGGATCAAAATAA... | 20 | taa | 1.00 | 0.00 | - | - | 1.00 |
| .......................................................................................................caatTGTGGGTACATGCTA........................................................... | 19 | caat | 1.00 | 0.00 | 1.00 | - | - |