| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (1) AGO2.ip | (1) B-CELL | (2) BRAIN | (32) BREAST | (16) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (3) HEART | (1) KIDNEY | (12) LIVER | (1) LUNG | (8) OTHER | (2) OVARY | (2) PLACENTA | (1) RRP40.ip | (8) SKIN | (2) SPLEEN | (4) UTERUS | (1) XRN.ip |
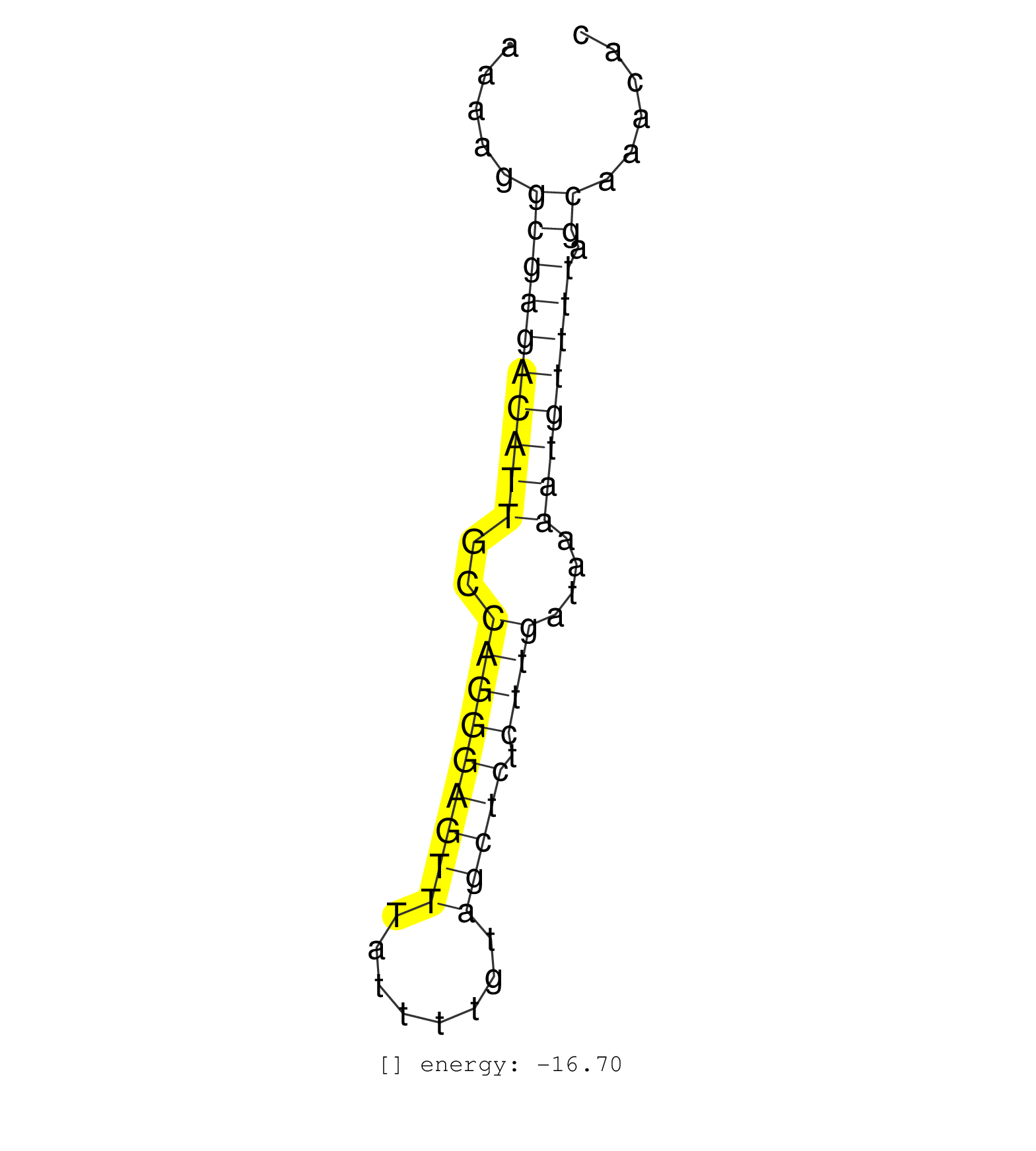
| AGAAATGTGGACTTTGCGGATGAAGTAGACCTCAATGTTGCCTTGCATCTAAAAGGCGAGACATTGCCAGGGAGTTTATTTTGTAGCTCTCTTGATAAAATGTTTTAGCAAACACTAGCGTGCCAGGCTTGGTCCTAAGCCCTTTGCCGGTGTTAACCCATGTAA .......................................................((((((((((..(((((((((........))))).))))....)))))))).))........................................................ ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577738(Rovira) total RNA. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577589(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR139195(SRX050645) FLASHPage purified small RNA (~15-40nt) from . (ovary) | TAX577739(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577746(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577744(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR343334 | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR029127(GSM416756) A549. (cell line) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR139197(SRX050645) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | TAX577590(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR191619(GSM715729) 166genomic small RNA (size selected RNA from . (breast) | SRR029131(GSM416760) MCF7. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR191412(GSM715522) 24genomic small RNA (size selected RNA from t. (breast) | SRR191451(GSM715561) 177genomic small RNA (size selected RNA from . (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | TAX577588(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR343336 | SRR191569(GSM715679) 54genomic small RNA (size selected RNA from t. (breast) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR191408(GSM715518) 88genomic small RNA (size selected RNA from t. (breast) | SRR191589(GSM715699) 69genomic small RNA (size selected RNA from t. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR191466(GSM715576) 114genomic small RNA (size selected RNA from . (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR189785 | SRR191570(GSM715680) 56genomic small RNA (size selected RNA from t. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR040038(GSM532923) G531N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR191618(GSM715728) 148genomic small RNA (size selected RNA from . (breast) | SRR343335 | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR191558(GSM715668) 61genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR191521(GSM715631) 92genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR343337 | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................ACATTGCCAGGGAGTTcca...................................................................................... | 19 | CCA | 132.00 | 2.00 | 47.00 | 9.00 | 3.00 | 4.00 | 3.00 | 3.00 | 6.00 | 3.00 | 1.00 | 5.00 | 2.00 | 4.00 | 2.00 | 1.00 | 1.00 | 2.00 | 2.00 | - | 2.00 | 3.00 | 1.00 | - | - | 1.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | - | 2.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcc....................................................................................... | 18 | CC | 59.00 | 2.00 | 16.00 | 1.00 | 2.00 | 4.00 | 1.00 | - | - | - | 2.00 | 1.00 | 1.00 | - | 2.00 | - | 1.00 | 2.00 | 2.00 | 2.00 | - | - | 1.00 | 2.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTacca...................................................................................... | 19 | ACCA | 44.00 | 0.00 | 5.00 | 3.00 | 7.00 | - | 2.00 | 3.00 | 1.00 | - | 3.00 | - | 3.00 | 4.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTc........................................................................................ | 17 | C | 30.00 | 2.00 | - | 4.00 | - | 1.00 | 1.00 | 5.00 | 3.00 | 2.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccaa..................................................................................... | 20 | CCAA | 29.00 | 2.00 | 12.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTacc....................................................................................... | 18 | ACC | 22.00 | 0.00 | 5.00 | 3.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccc...................................................................................... | 19 | CCC | 10.00 | 2.00 | 3.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcta...................................................................................... | 19 | CTA | 9.00 | 2.00 | 4.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTca....................................................................................... | 18 | CA | 9.00 | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccat..................................................................................... | 20 | CCAT | 9.00 | 2.00 | 3.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTTc....................................................................................... | 18 | C | 8.00 | 7.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTT........................................................................................ | 17 | 1 | 7.00 | 7.00 | - | 2.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTct....................................................................................... | 18 | CT | 6.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTac........................................................................................ | 17 | AC | 5.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccac..................................................................................... | 20 | CCAC | 5.00 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTatca...................................................................................... | 19 | ATCA | 5.00 | 0.00 | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTa........................................................................................ | 17 | A | 5.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................ACATTGCCAGGGAGTTTca...................................................................................... | 19 | CA | 5.00 | 7.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcaa...................................................................................... | 19 | CAA | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcct...................................................................................... | 19 | CCT | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GACATTGCCAGGGAGTacca...................................................................................... | 20 | ACCA | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTgcca...................................................................................... | 19 | GCCA | 3.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTacaa...................................................................................... | 19 | ACAA | 3.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGCCAGGGAGTTcca...................................................................................... | 18 | CCA | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTaca....................................................................................... | 18 | ACA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTataa...................................................................................... | 19 | ATAA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTTtaa..................................................................................... | 20 | TAA | 2.00 | 7.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccg...................................................................................... | 19 | CCG | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccag..................................................................................... | 20 | CCAG | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTctt...................................................................................... | 19 | CTT | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTctac..................................................................................... | 20 | CTAC | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAGACATTGCCAGGGAtttc........................................................................................ | 20 | TTTC | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTatc....................................................................................... | 18 | ATC | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTT......................................................................................... | 16 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccca..................................................................................... | 20 | CCCA | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTctaa..................................................................................... | 20 | CTAA | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTaccc...................................................................................... | 19 | ACCC | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GACATTGCCAGGGAGTTccta..................................................................................... | 21 | CCTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTTt....................................................................................... | 18 | T | 1.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcg....................................................................................... | 18 | CG | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTatcc...................................................................................... | 19 | ATCC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTccc....................................................................................... | 18 | CCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTTcct..................................................................................... | 20 | CCT | 1.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTacat...................................................................................... | 19 | ACAT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CTTTGCGGATGAAGTAGACCTCtg.................................................................................................................................. | 24 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CTTGATAAAATGTTTTtat........................................................ | 19 | TAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................GAGACATTGCCAGGGAtctg........................................................................................ | 20 | TCTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTctag..................................................................................... | 20 | CTAG | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcag...................................................................................... | 19 | CAG | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTacga...................................................................................... | 19 | ACGA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AGACATTGCCAGGGAatc......................................................................................... | 18 | ATC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAGACATTGCCAGGGAtttg........................................................................................ | 20 | TTTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTact....................................................................................... | 18 | ACT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTgca...................................................................................... | 19 | GCA | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTatat...................................................................................... | 19 | ATAT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AGACATTGCCAGGGAttac........................................................................................ | 19 | TTAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................AGGGAGTTTATTTTGggcc.............................................................................. | 19 | GGCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAGACATTGCCAGGGAtta......................................................................................... | 19 | TTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ATGTGGACTTTGCGGATGA.............................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTATTTTGTAGCTCTaatg....................................................................... | 19 | AATG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcccc..................................................................................... | 20 | CCCC | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTatct...................................................................................... | 19 | ATCT | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGCCAGGGAGTTct....................................................................................... | 17 | CT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTggt...................................................................................... | 19 | GGT | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTaggc...................................................................................... | 19 | AGGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AGACATTGCCAGGGAtttc........................................................................................ | 19 | TTTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTctca..................................................................................... | 20 | CTCA | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTccca...................................................................................... | 19 | CCCA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTccga..................................................................................... | 20 | CCGA | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ACATTGCCAGGGAGTTcctc..................................................................................... | 20 | CCTC | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAAATGTGGACTTTGCGGATGAAGTAGACCTCAATGTTGCCTTGCATCTAAAAGGCGAGACATTGCCAGGGAGTTTATTTTGTAGCTCTCTTGATAAAATGTTTTAGCAAACACTAGCGTGCCAGGCTTGGTCCTAAGCCCTTTGCCGGTGTTAACCCATGTAA .......................................................((((((((((..(((((((((........))))).))))....)))))))).))........................................................ ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577738(Rovira) total RNA. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577589(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR139195(SRX050645) FLASHPage purified small RNA (~15-40nt) from . (ovary) | TAX577739(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577746(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577744(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR343334 | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR029127(GSM416756) A549. (cell line) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR139197(SRX050645) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | TAX577590(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR191619(GSM715729) 166genomic small RNA (size selected RNA from . (breast) | SRR029131(GSM416760) MCF7. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR191412(GSM715522) 24genomic small RNA (size selected RNA from t. (breast) | SRR191451(GSM715561) 177genomic small RNA (size selected RNA from . (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | TAX577588(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR343336 | SRR191569(GSM715679) 54genomic small RNA (size selected RNA from t. (breast) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR191408(GSM715518) 88genomic small RNA (size selected RNA from t. (breast) | SRR191589(GSM715699) 69genomic small RNA (size selected RNA from t. (breast) | TAX577740(Rovira) total RNA. (breast) | SRR191466(GSM715576) 114genomic small RNA (size selected RNA from . (breast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR189785 | SRR191570(GSM715680) 56genomic small RNA (size selected RNA from t. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR040038(GSM532923) G531N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR191618(GSM715728) 148genomic small RNA (size selected RNA from . (breast) | SRR343335 | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR191558(GSM715668) 61genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR191521(GSM715631) 92genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR343337 | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................CAAACACTAGCGTGCC......................................... | 16 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................tccaCAAACACTAGCGTGCC......................................... | 20 | tcca | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ccaCAAACACTAGCGTGCC......................................... | 19 | cca | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |