| (3) BRAIN | (1) BREAST | (4) CELL-LINE | (2) CERVIX | (1) HEART | (1) OTHER | (1) SPLEEN |
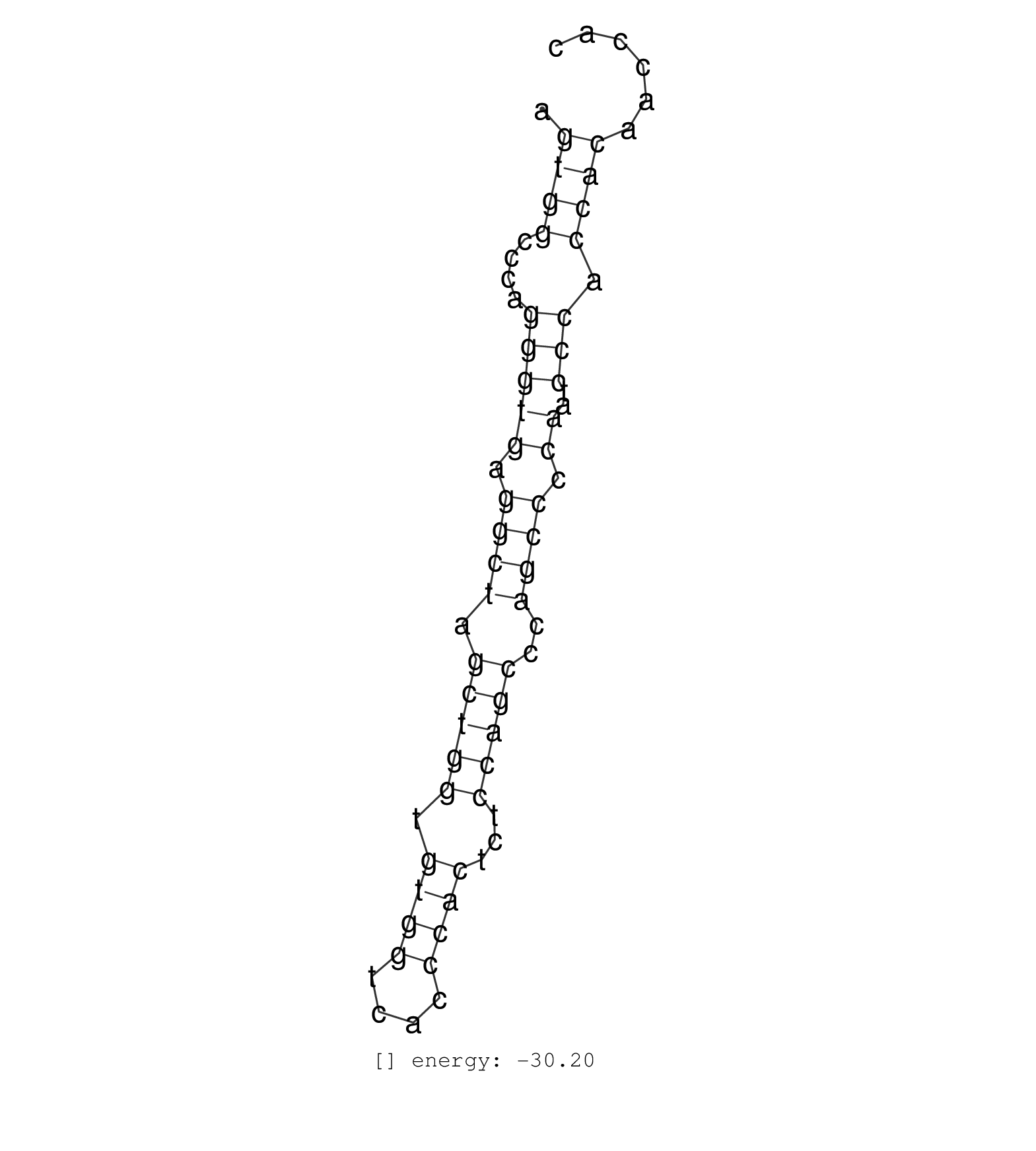
| CCAGGACACTCACAACGAGACCCCTGGGAAGATGAAACCCCTGGCTGCACAGTGGCCCAGGGTGAGGCTAGCTGGTGTGGTCACCCACTCTCCAGCCCAGCCCCAATCCCACCACAACCACATTCGACAAATGGCCCACTGTGGGCCAACGGACACGTGGCCATTCCAGAG ...................................................((((....(((((.((((.(((((.((((....))))...)))))..)))).))..))).))))........................................................ ..................................................51....................................................................121................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM532880(GSM532880) G659T. (cervix) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM532887(GSM532887) G761N. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................ATCCCACCACAACCACAa................................................ | 18 | A | 2.00 | 0.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................ATCCCACCACAACCACtat............................................... | 19 | TAT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TAGCTGGTGTGGTCACaatg................................................................................... | 20 | AATG | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGGTCACCCACTCTCCcgc........................................................................... | 19 | CGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| CCAGGACACTCACAACGAGACCCCTGGGAAGATGAAACCCCTGGCTGCACAGTGGCCCAGGGTGAGGCTAGCTGGTGTGGTCACCCACTCTCCAGCCCAGCCCCAATCCCACCACAACCACATTCGACAAATGGCCCACTGTGGGCCAACGGACACGTGGCCATTCCAGAG ...................................................((((....(((((.((((.(((((.((((....))))...)))))..)))).))..))).))))........................................................ ..................................................51....................................................................121................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM532880(GSM532880) G659T. (cervix) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM532887(GSM532887) G761N. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................ccctTCCCACCACAACCAC.................................................. | 19 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................taaaACCCACTCTCCAGCCCAG....................................................................... | 22 | taaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ccaaCCCAGCCCCAATCCCA............................................................ | 20 | ccaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................aACCCACTCTCCAGCCCAG....................................................................... | 19 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................gctgCTCCAGCCCAGCCCC................................................................... | 19 | gctg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................actcACCCCTGGCTGCACA........................................................................................................................ | 19 | actc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................CAAATGGCCCACTGTGGGCCAACGG................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................taatCCCTGGCTGCACAGT...................................................................................................................... | 19 | taat | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................tggcGCCCAGGGTGAGGCT...................................................................................................... | 19 | tggc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CTCCAGCCCAGCCCCAA................................................................. | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |