| (2) BRAIN | (2) BREAST | (2) CELL-LINE | (3) CERVIX | (1) FIBROBLAST | (1) LUNG | (2) OTHER | (1) SKIN |
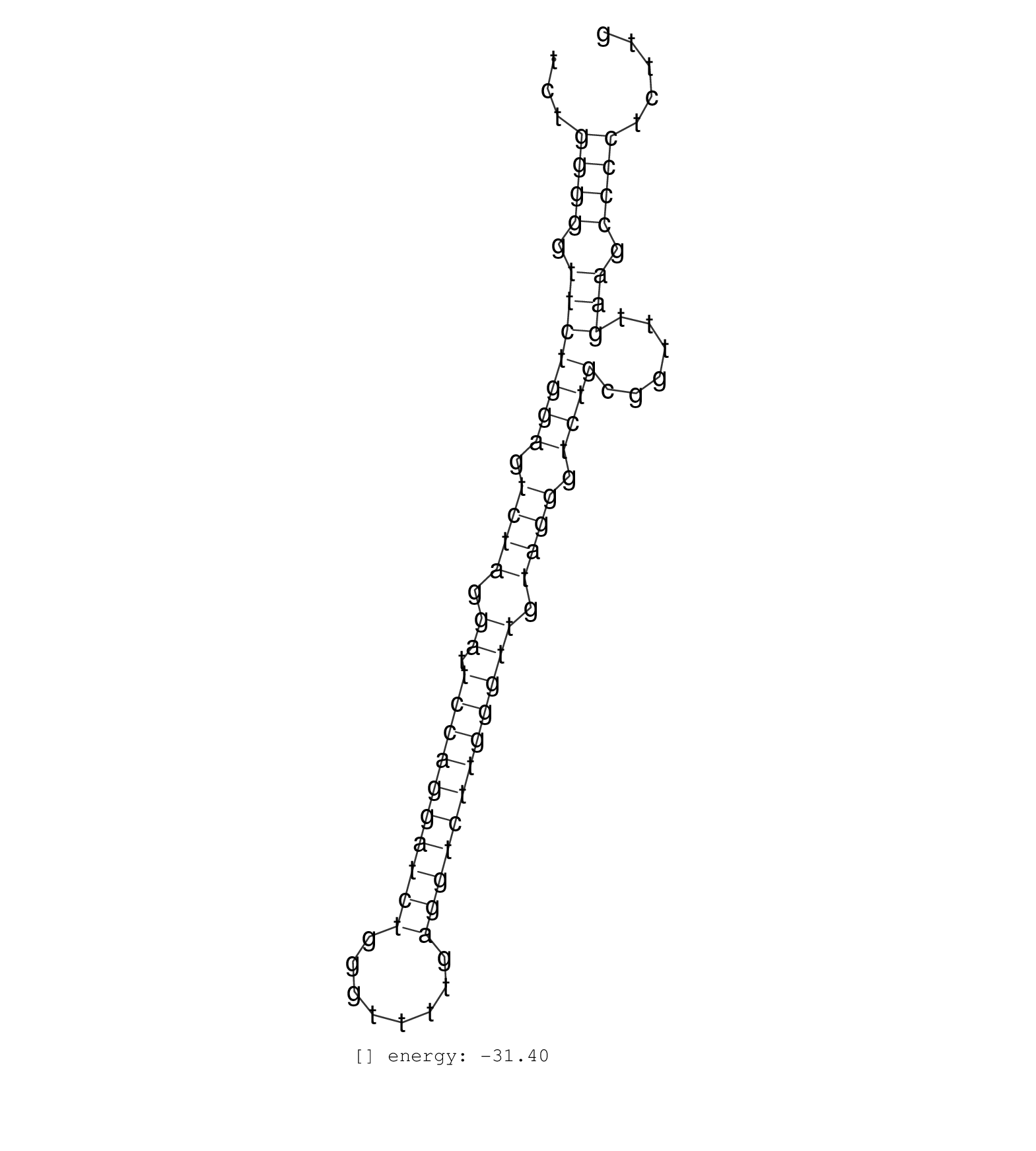
| CTACGTGGCCCCTGAGCAGCTCTGGGGAACATTCTTGGTCCGGGTTCCTGTCTGGGGGTTCTGGAGTCTAGGATTCCAGGATCTGGGTTTTGAGGTCTTGGGTTGTAGGGTCTGCGGTTTGAAGCCCCTCTTGTAACCCAGGACAGAGGTGAAAGCAGTGGGGTGGACTGGGCAGCCCCCAAA .....................................................((((.(((((((.((((.((.((((((((((........)))))))))))).)))).))))......))).))))....................................................... ..................................................51................................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR040031(GSM532916) G013T. (cervix) | SRR040008(GSM532893) G727N. (cervix) | TAX577742(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR189784 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................ATCTGGGTTTTGAGGact..................................................................................... | 18 | ACT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................CTGGGGGTTCTGGAGacg.................................................................................................................. | 18 | ACG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTAGaggg....................................................................... | 22 | AGGG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTgttt......................................................................... | 20 | GTTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTAGatc........................................................................ | 21 | ATC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTActgt........................................................................ | 21 | CTGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................TGGGGGTTCTGGAGTCTggtg.............................................................................................................. | 21 | GGTG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTAtagt........................................................................ | 21 | TAGT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGGGGGTTCTGGAGTCTgtg............................................................................................................... | 20 | GTG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAGGTCTTGGGTTGTgtt.......................................................................... | 19 | GTT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............AGCAGCTCTGGGGAAaatg...................................................................................................................................................... | 19 | AATG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTGAAGCCCCTCTTG.................................................. | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| CTACGTGGCCCCTGAGCAGCTCTGGGGAACATTCTTGGTCCGGGTTCCTGTCTGGGGGTTCTGGAGTCTAGGATTCCAGGATCTGGGTTTTGAGGTCTTGGGTTGTAGGGTCTGCGGTTTGAAGCCCCTCTTGTAACCCAGGACAGAGGTGAAAGCAGTGGGGTGGACTGGGCAGCCCCCAAA .....................................................((((.(((((((.((((.((.((((((((((........)))))))))))).)))).))))......))).))))....................................................... ..................................................51................................................................................133................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR040031(GSM532916) G013T. (cervix) | SRR040008(GSM532893) G727N. (cervix) | TAX577742(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR189784 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................tgcTTCCTGTCTGGGGGT............................................................................................................................ | 18 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................gtCTGTCTGGGGGTTCTG........................................................................................................................ | 18 | gt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCCTCTTGTAACCCAGGACAGA.................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |