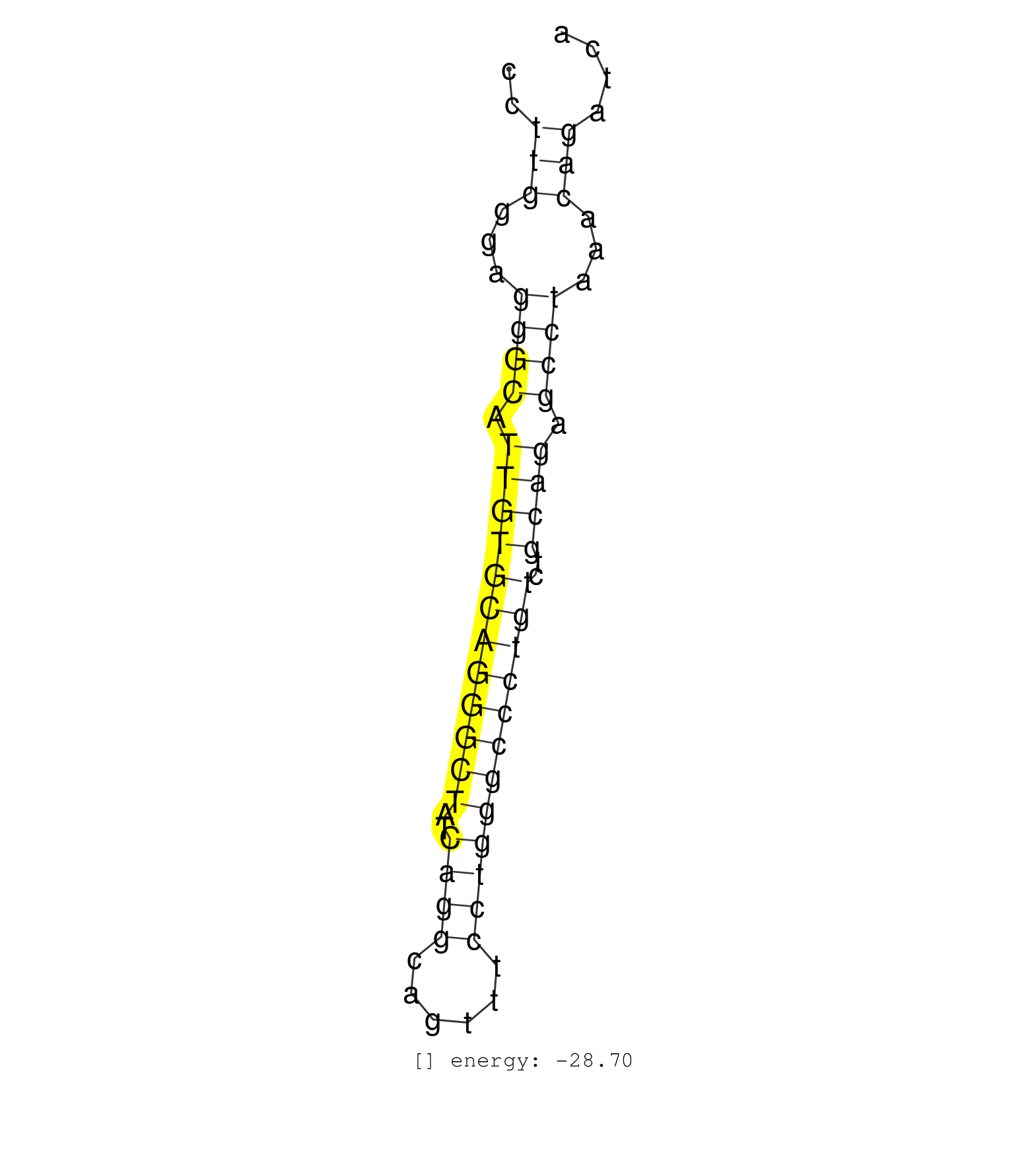
| (1) AGO2.ip | (1) BRAIN | (3) BREAST | (25) CELL-LINE | (2) CERVIX | (1) HELA | (2) OTHER | (5) SKIN |
| AGCATGGTATCTCTGCCCTAGATGCACTGATGGCAGCAGAGAGCTCTGTCCCTTGGGAGGGCATTGTGCAGGGCTATCAGGCAGTTTCCTGGGCCCTGTCTGCAGAGCCTAAACAGATCACATCTGGCCAGTCTCAACAGGGCCTCCTTACCCTATAAACCATGCATGTA ....................................................(((...((((.((((((((((((..((((......))))))))))))..)))).))))...)))...................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR189782 | SRR040006(GSM532891) G601N. (cervix) | SRR029124(GSM416753) HeLa. (hela) | SRR029129(GSM416758) SW480. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR037935(GSM510473) 293cand3. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR444055(SRX128903) Sample 27_2cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR191495(GSM715605) 159genomic small RNA (size selected RNA from . (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GCATTGTGCAGGGCTATga........................................................................................... | 19 | GA | 55.00 | 0.00 | 4.00 | 4.00 | 1.00 | 4.00 | 3.00 | 2.00 | 4.00 | 4.00 | 2.00 | 2.00 | 3.00 | 3.00 | - | - | 2.00 | 1.00 | 1.00 | 1.00 | - | 3.00 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATga........................................................................................... | 18 | GA | 17.00 | 1.00 | - | 9.00 | - | - | 1.00 | - | - | 1.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATg............................................................................................ | 18 | G | 15.00 | 0.00 | 5.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgat.......................................................................................... | 20 | GAT | 13.00 | 0.00 | - | - | - | 2.00 | 2.00 | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATC............................................................................................ | 18 | 1 | 10.00 | 10.00 | - | - | 8.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgt........................................................................................... | 19 | GT | 9.00 | 0.00 | - | - | 1.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATga........................................................................................... | 17 | GA | 5.40 | 0.20 | - | 4.60 | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | 0.20 | 0.20 |
| .............................................................CATTGTGCAGGGCTATgat.......................................................................................... | 19 | GAT | 4.00 | 1.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgaa.......................................................................................... | 20 | GAA | 4.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATgt........................................................................................... | 18 | GT | 3.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATCt........................................................................................... | 19 | T | 2.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgg........................................................................................... | 19 | GG | 2.00 | 0.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................GTCCCTTGGGAGGGCgcg......................................................................................................... | 18 | GCG | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATg............................................................................................ | 17 | G | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgga.......................................................................................... | 20 | GGA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgc........................................................................................... | 19 | GC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATCA........................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTATAAACCATGCATttc | 18 | TTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgatt......................................................................................... | 21 | GATT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTgtga........................................................................................... | 19 | GTGA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATC............................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTgtc............................................................................................ | 18 | GTC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATCA........................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTgtg............................................................................................ | 18 | GTG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATaa........................................................................................... | 19 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTATgaa.......................................................................................... | 19 | GAA | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTATgta.......................................................................................... | 20 | GTA | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GCATTGTGCAGGGCTctga........................................................................................... | 19 | CTGA | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CATTGTGCAGGGCTAT............................................................................................. | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATgat.......................................................................................... | 18 | GAT | 0.80 | 0.20 | - | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATg............................................................................................ | 16 | G | 0.80 | 0.20 | - | 0.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATCA........................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATaa........................................................................................... | 17 | AA | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| ..............................................................ATTGTGCAGGGCTAT............................................................................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ..............................................................ATTGTGCAGGGCTATgag.......................................................................................... | 18 | GAG | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATgaa.......................................................................................... | 18 | GAA | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTGTGCAGGGCTATgaag......................................................................................... | 19 | GAAG | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCATGGTATCTCTGCCCTAGATGCACTGATGGCAGCAGAGAGCTCTGTCCCTTGGGAGGGCATTGTGCAGGGCTATCAGGCAGTTTCCTGGGCCCTGTCTGCAGAGCCTAAACAGATCACATCTGGCCAGTCTCAACAGGGCCTCCTTACCCTATAAACCATGCATGTA ....................................................(((...((((.((((((((((((..((((......))))))))))))..)))).))))...)))...................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR189782 | SRR040006(GSM532891) G601N. (cervix) | SRR029124(GSM416753) HeLa. (hela) | SRR029129(GSM416758) SW480. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR037935(GSM510473) 293cand3. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR029127(GSM416756) A549. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR444055(SRX128903) Sample 27_2cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR191495(GSM715605) 159genomic small RNA (size selected RNA from . (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........taTCTGCCCTAGATGCAC............................................................................................................................................... | 18 | ta | 16.00 | 0.00 | 7.00 | - | - | - | 2.00 | 1.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......tctaTCTGCCCTAGATGCAC............................................................................................................................................... | 20 | tcta | 9.00 | 0.00 | 3.00 | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ctaTCTGCCCTAGATGCAC............................................................................................................................................... | 19 | cta | 9.00 | 0.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........aTCTGCCCTAGATGCAC............................................................................................................................................... | 17 | a | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......tctaTCTGCCCTAGATGCA................................................................................................................................................ | 19 | tcta | 3.00 | 0.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......actaTCTGCCCTAGATGCA................................................................................................................................................ | 19 | acta | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......actaTCTGCCCTAGATGCAC............................................................................................................................................... | 20 | acta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCTCTGCCCTAGATGCAC............................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ctaTCTGCCCTAGATGCA................................................................................................................................................ | 18 | cta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......tataTCTGCCCTAGATGCAC............................................................................................................................................... | 20 | tata | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |