| (1) AGO2.ip | (1) B-CELL | (5) BREAST | (3) CELL-LINE | (1) HEART | (1) HELA | (1) PLACENTA | (1) SKIN |
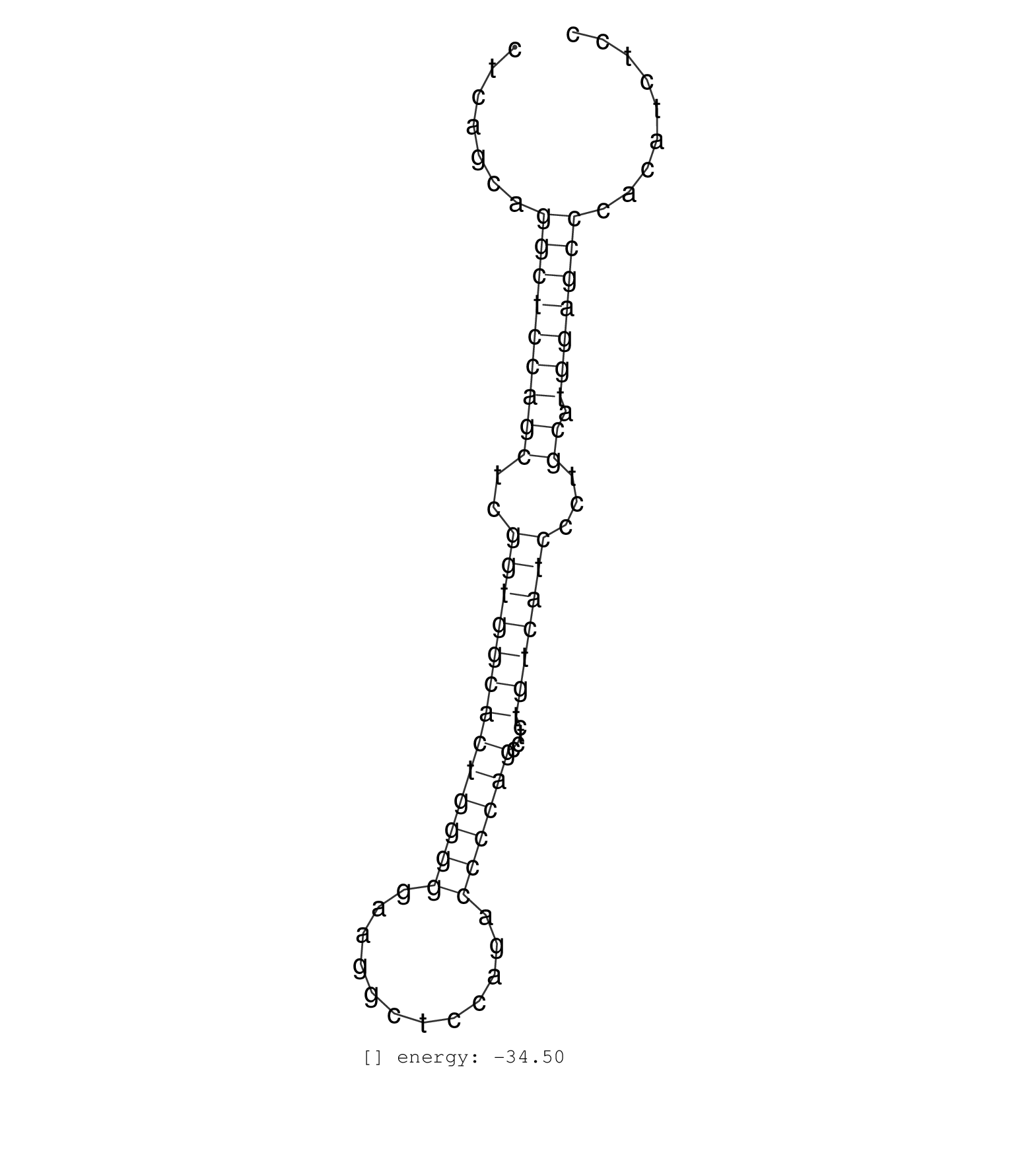
| AGGGGTGGTATCTCCGAAGGAGCCGAGGATTCTCCGTGATGACTTGCAAACTCAGCAGGCTCCAGCTCGGTGGCACTGGGGGAAGGCTCCAGACCCCAGCCTCTGTCATCCCTGCATGGAGCCCACATCTCCAAGGAGGCTGGGAGATCCTGTTTGGGGGTCAATTTAAATGTGAAGGAGCA .........................................................(((((((((..(((((((((((((............))))))....)))))))...)).)))))))........................................................... ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................GGAGGCTGGGAGATCCctt............................. | 19 | CTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GAGGCTGGGAGATCCTGc............................. | 18 | C | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCTGTCATCCCTGCATtt............................................................... | 18 | TT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GAGGCTGGGAGATCCctt............................. | 18 | CTT | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................CCCTGCATGGAGCCCgt........................................................ | 17 | GT | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .GGGGTGGTATCTCCGtgg................................................................................................................................................................... | 18 | TGG | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AAGGAGGCTGGGAGATCCTGTTTGGG........................ | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................................CCCACATCTCCAAGGAG............................................ | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| AGGGGTGGTATCTCCGAAGGAGCCGAGGATTCTCCGTGATGACTTGCAAACTCAGCAGGCTCCAGCTCGGTGGCACTGGGGGAAGGCTCCAGACCCCAGCCTCTGTCATCCCTGCATGGAGCCCACATCTCCAAGGAGGCTGGGAGATCCTGTTTGGGGGTCAATTTAAATGTGAAGGAGCA .........................................................(((((((((..(((((((((((((............))))))....)))))))...)).)))))))........................................................... ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................caGCTCCAGCTCGGTGGC............................................................................................................ | 18 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................ctagAGACCCCAGCCTCTGTC........................................................................... | 21 | ctag | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................ctagCCAGACCCCAGCCTC............................................................................... | 19 | ctag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................aaaaCCGTGATGACTTGCA...................................................................................................................................... | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........gccCCGAAGGAGCCGAGG.......................................................................................................................................................... | 18 | gcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |