| (1) B-CELL | (1) BRAIN | (2) BREAST | (4) CELL-LINE | (1) CERVIX | (3) OTHER | (2) SKIN | (2) SPLEEN |
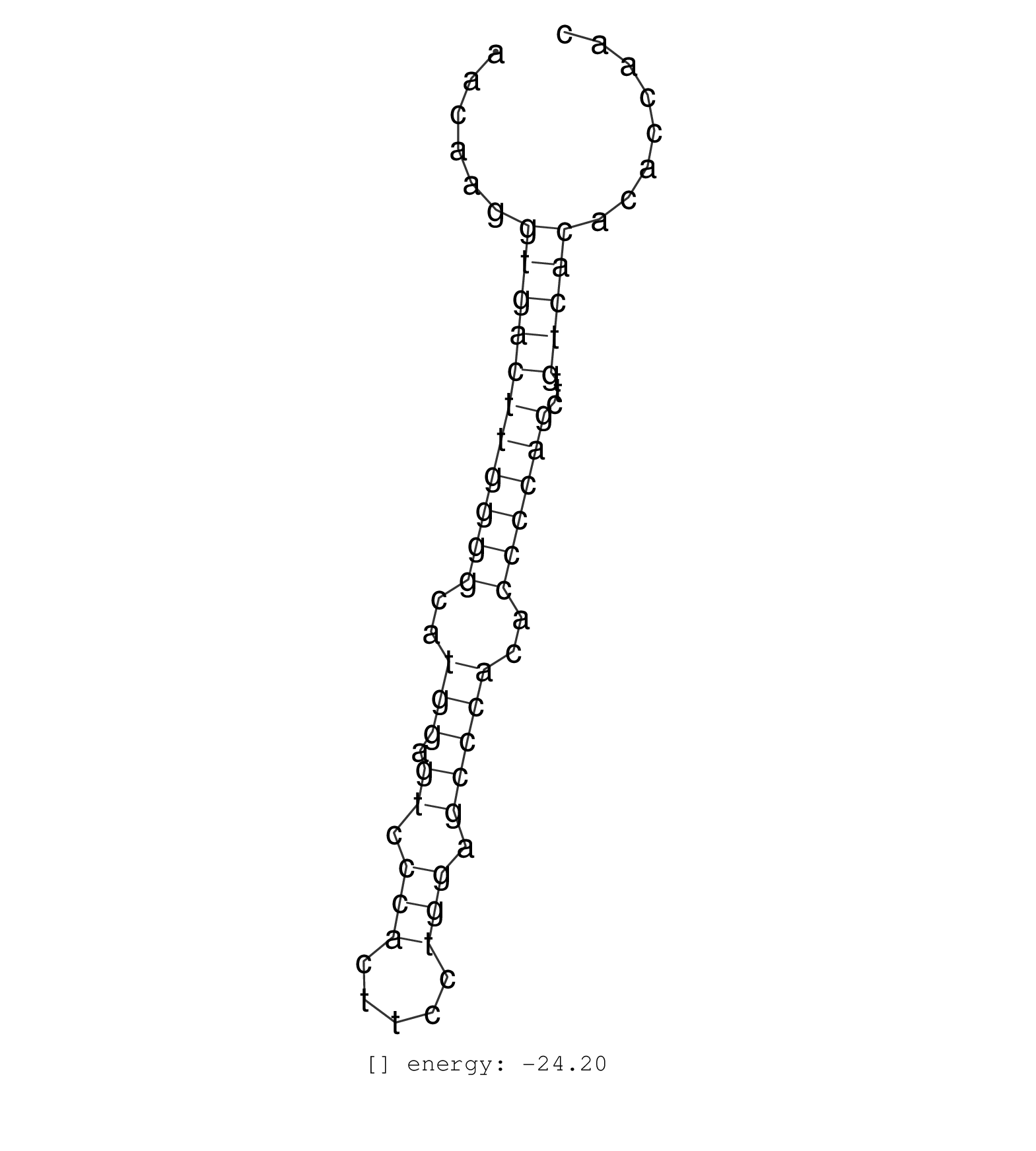
| TGGCCCTGAGGCTGGAGATGTCTTCGTTGCCTGACCTGACACCCACCTTCAACAAGGTGACTTGGGGCATGGAGTCCCACTTCCTGGAGCCCACACCCCAGCTTGTCACACACCAACTGCTGCCACCTGCTTCCTATTAGGGCAGATCTGCCCCTGTGAGAACCCCA ........................................................(((((((((((..(((.((.(((.....))).)))))..))))))...))))).......................................................... ..................................................51................................................................117................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | GSM532876(GSM532876) G547T. (cervix) | SRR189782 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR139205(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577453(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................ACACACCAACTGCTGCtttg........................................ | 20 | TTTG | 15.00 | 0.00 | 14.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................ACACACCAACTGCTGCttt......................................... | 19 | TTT | 5.00 | 0.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................ACACACCAACTGCTGCtctg........................................ | 20 | TCTG | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........AGGCTGGAGATGTCTTggg............................................................................................................................................ | 19 | GGG | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGAGGCTGGAGATGTCTTCGTT........................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .GGCCCTGAGGCTGGAGAT.................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTGAGGCTGGAGATGTCTTCG............................................................................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CACACCAACTGCTGCtcag........................................ | 19 | TCAG | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................ACACACCAACTGCTGCtt.......................................... | 18 | TT | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CTGGAGATGTCTTCGTTGCCTGACCTG................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......TGAGGCTGGAGATGTCTTCG............................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......TGAGGCTGGAGATGTatag.............................................................................................................................................. | 19 | ATAG | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................ACACACCAACTGCTGCtgtg........................................ | 20 | TGTG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GGCCCTGAGGCTGGAGATG................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCTTCGTTGCCTGACCTGACACCC........................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................GTCACACACCAACTGttg............................................. | 18 | TTG | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......TGAGGCTGGAGATGTCTTC.............................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................GACACCCACCTTCAACAAGct............................................................................................................. | 21 | CT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GACTTGGGGCATGGAccg........................................................................................... | 18 | CCG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TGGCCCTGAGGCTGGAGATGTCTTCGTTGCCTGACCTGACACCCACCTTCAACAAGGTGACTTGGGGCATGGAGTCCCACTTCCTGGAGCCCACACCCCAGCTTGTCACACACCAACTGCTGCCACCTGCTTCCTATTAGGGCAGATCTGCCCCTGTGAGAACCCCA ........................................................(((((((((((..(((.((.(((.....))).)))))..))))))...))))).......................................................... ..................................................51................................................................117................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189784 | GSM532876(GSM532876) G547T. (cervix) | SRR189782 | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR139205(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577453(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................acccCTGCTGCCACCTGCT.................................... | 19 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................ccgTTGGGGCATGGAGTC........................................................................................... | 18 | ccg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................agTTCGTTGCCTGACCTG................................................................................................................................. | 18 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................ccaTTGGGGCATGGAGTC........................................................................................... | 18 | cca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |