| (2) AGO2.ip | (5) B-CELL | (2) BRAIN | (9) BREAST | (51) CELL-LINE | (3) CERVIX | (1) FIBROBLAST | (1) HEART | (2) HELA | (2) KIDNEY | (7) LIVER | (2) LUNG | (1) OTHER | (1) RRP40.ip | (12) SKIN | (1) SPLEEN | (3) TESTES | (2) THYMUS | (3) UTERUS | (1) XRN.ip |
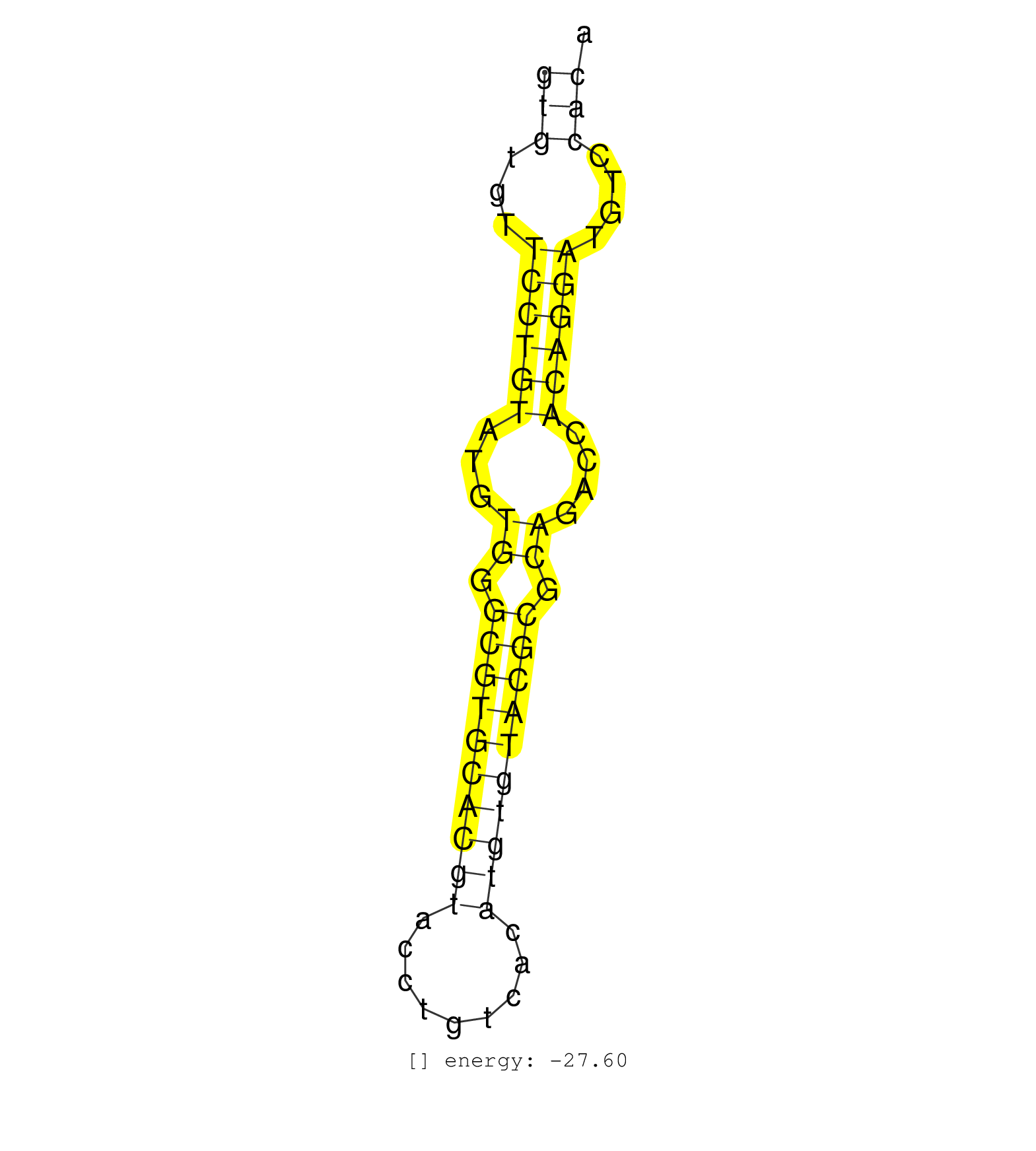
| TCTTAGGAGACGCCTGCCGGCCGCTTTGGGATAGGGCCTGTCTGCCCATGCTGGCTTCCAAAGGCCTCTGTGTGTTCCTGTATGTGGGCGTGCACGTACCTGTCACATGTGTACGCGCAGACCACAGGATGTCCACACTGGCTTCCAAACACATCTCTGTGTTTCTGTCTGTGAGTGTGTACGCAGGTGTCAAATGTGTACGCACA ..................................................(((...((((((...((.((((((((((.........)))))))))).))....))))))....)))................................................... .....................................................................70.................................................................137................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189786 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM416733(GSM416733) HEK293. (cell line) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | GSM339994(GSM339994) hues6. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR189787 | SRR029128(GSM416757) H520. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR189782 | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM339995(GSM339995) hues6NP. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR038859(GSM458542) MM386. (cell line) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR029129(GSM416758) SW480. (cell line) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR139186(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR040011(GSM532896) G529T. (cervix) | SRR189783 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040036(GSM532921) G243N. (cervix) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR038858(GSM458541) MEL202. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR029130(GSM416759) DLD2. (cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR040043(GSM532928) G428T. (cervix) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR191430(GSM715540) 167genomic small RNA (size selected RNA from . (breast) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TACGCGCAGACCACAGGATGTC......................................................................... | 22 | 1 | 116.00 | 116.00 | 13.00 | 13.00 | 8.00 | 8.00 | 1.00 | 4.00 | - | 5.00 | 6.00 | 7.00 | 2.00 | 9.00 | 1.00 | 2.00 | - | 2.00 | 2.00 | 3.00 | - | 3.00 | 1.00 | - | 5.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | - | 1.00 | 3.00 | - | 2.00 | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCt........................................................................ | 23 | T | 47.00 | 116.00 | 4.00 | 6.00 | 3.00 | 1.00 | 5.00 | 3.00 | - | 1.00 | 6.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGT.......................................................................... | 21 | 1 | 37.00 | 37.00 | 7.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCa........................................................................ | 23 | A | 33.00 | 116.00 | 1.00 | 2.00 | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAC............................................................................................................... | 21 | 1 | 25.00 | 25.00 | - | - | - | 4.00 | 1.00 | 3.00 | - | - | - | - | 3.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCA................................................................................................................ | 20 | 1 | 21.00 | 21.00 | - | - | 3.00 | 1.00 | - | 4.00 | - | - | - | 1.00 | 2.00 | - | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCC........................................................................ | 23 | 1 | 19.00 | 19.00 | 5.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATG........................................................................... | 20 | 1 | 12.00 | 12.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAa............................................................................................................... | 21 | A | 7.00 | 21.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCatc...................................................................... | 25 | ATC | 6.00 | 116.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGA............................................................................. | 18 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGC................................................................................................................. | 19 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAt............................................................................................................... | 21 | T | 5.00 | 21.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCACG.............................................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCat....................................................................... | 24 | AT | 4.00 | 116.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCACt.............................................................................................................. | 22 | T | 4.00 | 25.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCACtt............................................................................................................. | 23 | TT | 4.00 | 25.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTACGCGCAGACCACAGGATGT.......................................................................... | 23 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGAcgtc......................................................................... | 22 | CGTC | 3.00 | 5.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGcc......................................................................... | 22 | CC | 3.00 | 12.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCCTGTATGTGGGCGTGCA................................................................................................................ | 19 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATa........................................................................... | 20 | A | 3.00 | 0.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAatc............................................................................................................. | 23 | ATC | 2.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTt......................................................................... | 22 | T | 2.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCACta............................................................................................................. | 23 | TA | 2.00 | 25.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGccc........................................................................ | 23 | CCC | 2.00 | 12.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTACGCGCAGACCACAGGATG........................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGac......................................................................... | 22 | AC | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGtt................................................................................................................ | 20 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGcg............................................................................ | 19 | CG | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGagt........................................................................ | 23 | AGT | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTat........................................................................ | 23 | AT | 1.00 | 37.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGgc......................................................................... | 22 | GC | 1.00 | 12.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTACGCAGGTGTCAAtga.......... | 18 | TGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCACtttt........................................................................................................... | 25 | TTTT | 1.00 | 25.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCACATGTGTACGCGCAGACCACAGGAc............................................................................ | 28 | C | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGcct........................................................................ | 23 | CCT | 1.00 | 12.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGAggg.......................................................................... | 21 | GGG | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTCtaa...................................................................... | 25 | TAA | 1.00 | 116.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGT................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ACATCTCTGTGTTTCTGTCTt................................... | 21 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTta........................................................................ | 23 | TA | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ACTGGCTTCCAAACACATCgg................................................. | 21 | GG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATatca........................................................................ | 23 | ATCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTaa........................................................................ | 23 | AA | 1.00 | 37.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CGCGCAGACCACAGGATGT.......................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CGCAGGTGTCAAATGTGTACGCA.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAtg.............................................................................................................. | 22 | TG | 1.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGG.............................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGTGTTCCTGTATttc........................................................................................................................ | 18 | TTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTACGCAGGTGTCAAcga.......... | 18 | CGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTACGCAGGTGTCAAcgt.......... | 18 | CGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTccac............................................................................................................... | 21 | CCAC | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTtc........................................................................ | 23 | TC | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................ACGCGCAGACCACAGGATGTCC........................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGgca........................................................................ | 23 | GCA | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATttcc........................................................................ | 23 | TTCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTACGCGCAGACCACAGGAcgtc......................................................................... | 24 | CGTC | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTACGCGCAGACCACAGGAc............................................................................ | 21 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGATGTtt........................................................................ | 23 | TT | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCCTGTATGTGGGCGTGCAaa.............................................................................................................. | 22 | AA | 1.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGTACGCGCAGACCACAGGATG........................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGAggtc......................................................................... | 22 | GGTC | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TACGCGCAGACCACAGGAcgac......................................................................... | 22 | CGAC | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTTAGGAGACGCCTGCCGGCCGCTTTGGGATAGGGCCTGTCTGCCCATGCTGGCTTCCAAAGGCCTCTGTGTGTTCCTGTATGTGGGCGTGCACGTACCTGTCACATGTGTACGCGCAGACCACAGGATGTCCACACTGGCTTCCAAACACATCTCTGTGTTTCTGTCTGTGAGTGTGTACGCAGGTGTCAAATGTGTACGCACA ..................................................(((...((((((...((.((((((((((.........)))))))))).))....))))))....)))................................................... .....................................................................70.................................................................137................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189786 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM416733(GSM416733) HEK293. (cell line) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | GSM339994(GSM339994) hues6. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR189787 | SRR029128(GSM416757) H520. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR189782 | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR029131(GSM416760) MCF7. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM339995(GSM339995) hues6NP. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR038859(GSM458542) MM386. (cell line) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR029129(GSM416758) SW480. (cell line) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR139186(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR040011(GSM532896) G529T. (cervix) | SRR189783 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040036(GSM532921) G243N. (cervix) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR038858(GSM458541) MEL202. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139207(SRX050653) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR029130(GSM416759) DLD2. (cell line) | SRR038860(GSM458543) MM426. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR038857(GSM458540) D20. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM359191(GSM359191) HepG2_3pM_5. (cell line) | SRR040043(GSM532928) G428T. (cervix) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR191430(GSM715540) 167genomic small RNA (size selected RNA from . (breast) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........GCCTGCCGGCCGCTTTGGG................................................................................................................................................................................ | 19 | 1 | 4.00 | 4.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................cccGTCTGCCCATGCTGG........................................................................................................................................................ | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................gcaCAGACCACAGGATGTCCA....................................................................... | 21 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CGCCTGCCGGCCGCTTTGGGAT.............................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................aATGTCCACACTGGCTTCCAAA......................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................ATGTCCACACTGGCTTCCAAACACATC................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................tCATGCTGGCTTCCAAAGGCC............................................................................................................................................ | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................tCTGGCTTCCAAACACAT.................................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |