| (2) BREAST | (2) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (1) HEART | (2) SKIN | (1) SPLEEN | (1) THYMUS | (1) UTERUS |
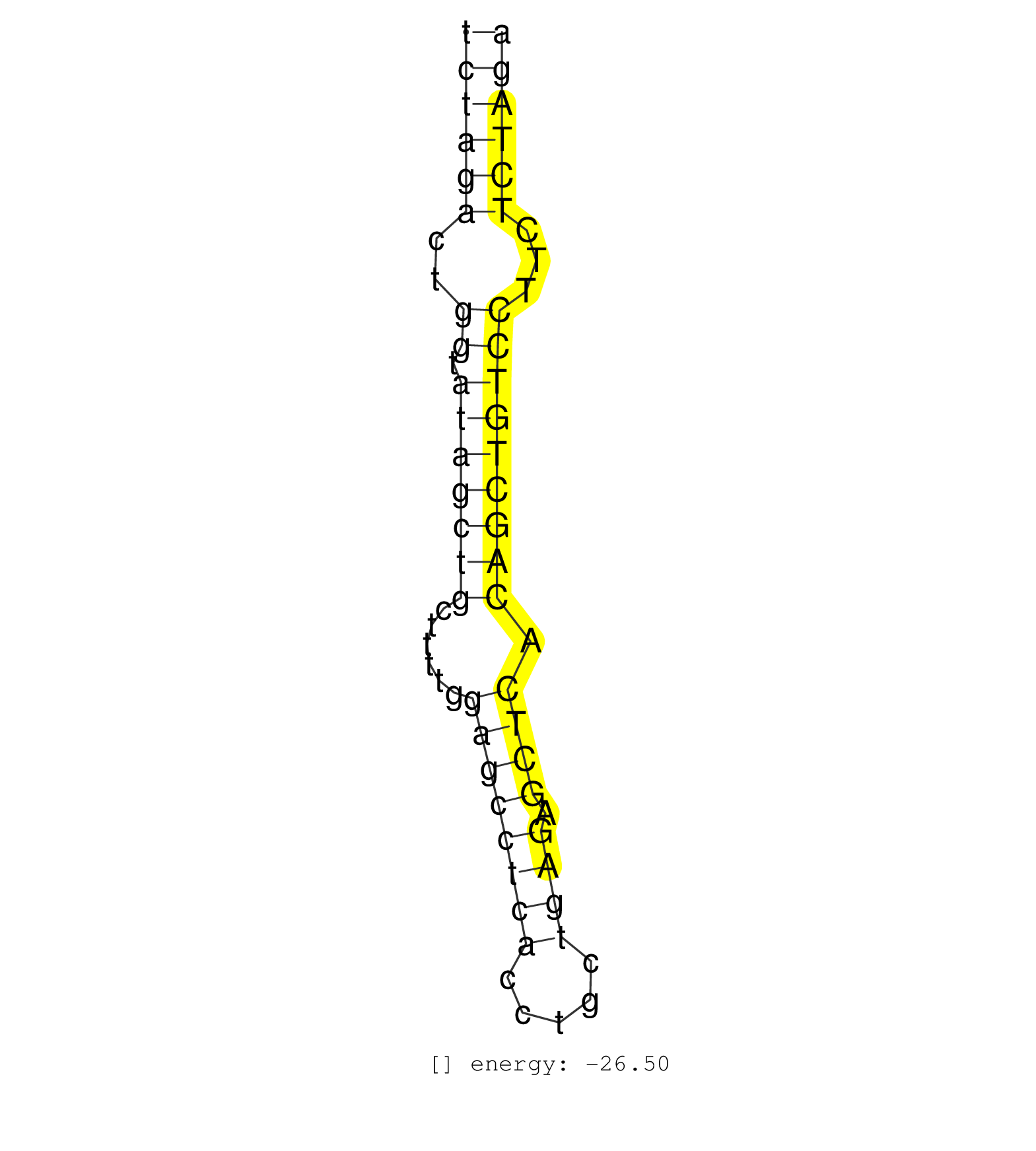
| TGTTATGAAAATCATCCCTGCCTAGAGATGAATTCCCCCTTCCCCTGAGGTCTAGACTGGTATAGCTGCTTTTGGAGCCTCACCTGCTGAGAGCTCACAGCTGTCCTTCTCTAGAGAATCACCCTCAGATGGGAGCCACATTGCCTGGGATGGGATGCCATGCCC ..................................................((((((..((.(((((((......((((((((.....)))).)))).)))))))))...)))))).................................................. ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR040029(GSM532914) G026T. (cervix) | TAX577740(Rovira) total RNA. (breast) | SRR040008(GSM532893) G727N. (cervix) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040018(GSM532903) G701N. (cervix) | SRR040040(GSM532925) G612N. (cervix) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CACATTGCCTGGGATttcc.......... | 19 | TTCC | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................ACATTGCCTGGGATGtcc.......... | 18 | TCC | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................GCCACATTGCCTGGGttt............. | 18 | TTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCACATTGCCTGGGATtaga.......... | 20 | TAGA | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GCCACATTGCCTGGGtttt............ | 19 | TTTT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................TCCCCTGAGGTCTAGtggt.......................................................................................................... | 19 | TGGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................AGCCACATTGCCTGGcggc............. | 19 | CGGC | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACATTGCCTGGGATtacc.......... | 19 | TACC | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CACATTGCCTGGGATttc........... | 18 | TTC | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GCCACATTGCCTGGGATttgt.......... | 21 | TTGT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TCCCCTGAGGTCTAGcgcc.......................................................................................................... | 19 | CGCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................AGAGCTCACAGCTGTCCTTCTCTA.................................................... | 24 | 4 | 0.75 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - |
| ..................................................................................................................AGAATCACCCTCAGAT................................... | 16 | 9 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 |
| TGTTATGAAAATCATCCCTGCCTAGAGATGAATTCCCCCTTCCCCTGAGGTCTAGACTGGTATAGCTGCTTTTGGAGCCTCACCTGCTGAGAGCTCACAGCTGTCCTTCTCTAGAGAATCACCCTCAGATGGGAGCCACATTGCCTGGGATGGGATGCCATGCCC ..................................................((((((..((.(((((((......((((((((.....)))).)))).)))))))))...)))))).................................................. ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR040029(GSM532914) G026T. (cervix) | TAX577740(Rovira) total RNA. (breast) | SRR040008(GSM532893) G727N. (cervix) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040018(GSM532903) G701N. (cervix) | SRR040040(GSM532925) G612N. (cervix) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................TGAATTCCCCCTTCCCCTGA..................................................................................................................... | 20 | 4 | 0.75 | 0.75 | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................ATGAATTCCCCCTTCCCCTGAGGTCTAG.............................................................................................................. | 28 | 4 | 0.75 | 0.75 | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGCTGTCCTTCTCTAGA.................................................. | 17 | 5 | 0.60 | 0.60 | - | - | - | - | - | - | - | - | - | - | - | - | 0.60 | - | - |