| (3) B-CELL | (3) CELL-LINE | (1) CERVIX | (1) HEART | (1) OTHER |
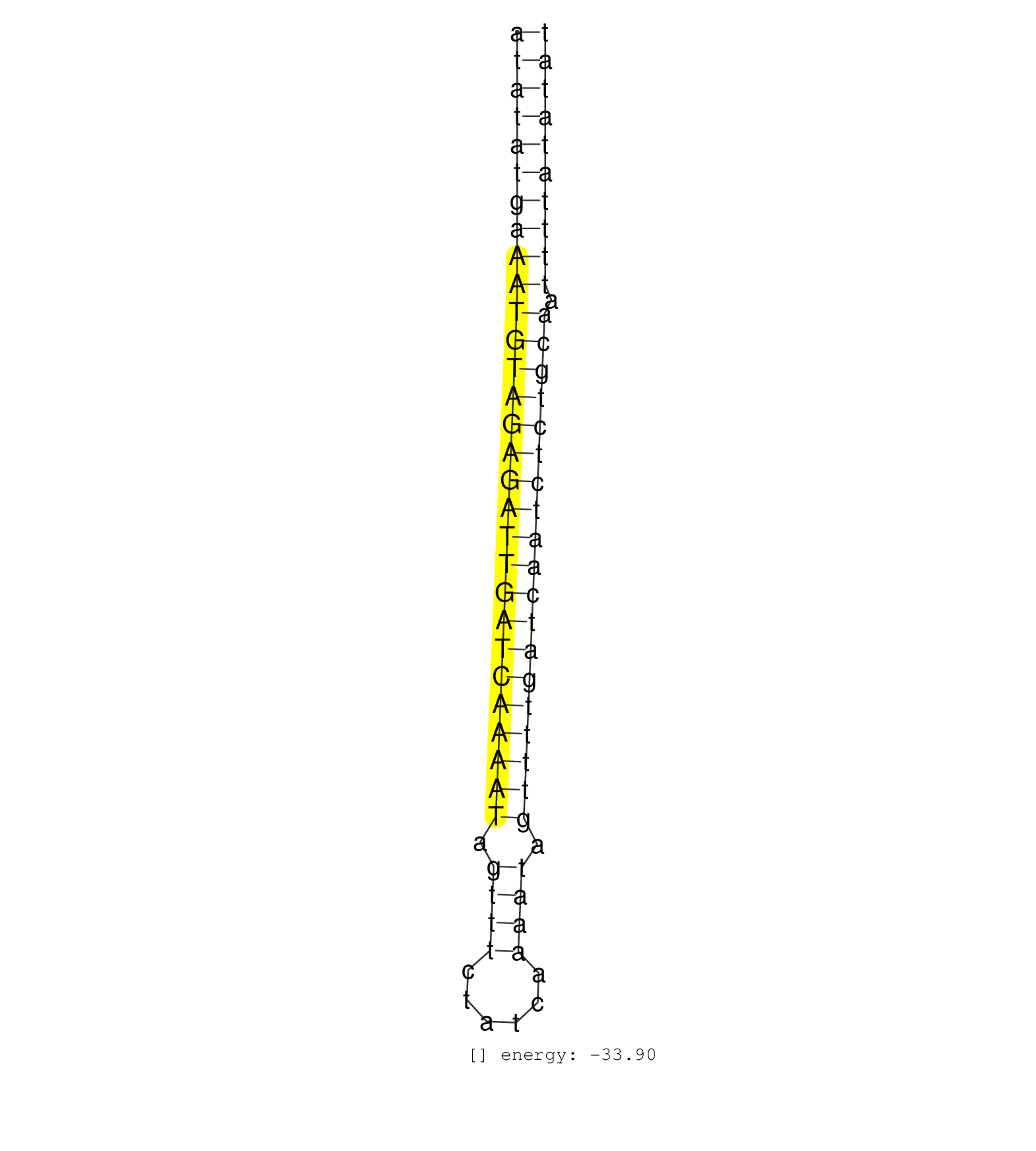
| TAACTATTTTACAACTATTTAAAAATGTCTTAGAGTTTAACTGGCCTAAAATATATGAAATGTAGAGATTGATCAAAATAGTTTCTATCAAAATAGTTTTGATCAATCTCTGCAATTTTATATATGAGGAAACTGAAGTCTGAAAGTATTATGCCCTTTGTGTCTCTATTTTGAA ..................................................(((((((((((((((((((((((((((((.((((......)))).))))))))))))))))))).)))))))))).................................................. ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR343336 | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR040012(GSM532897) G648N. (cervix) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR343337 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................AATGTAGAGATTGATCAAAAT................................................................................................ | 21 | 1 | 37.00 | 37.00 | 12.00 | 10.00 | 10.00 | 4.00 | - | 1.00 | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAATA............................................................................................... | 22 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - |
| ...........................................................ATGTAGAGATTGATCcct.................................................................................................. | 18 | CCT | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .........................................................AAATGTAGAGATTGATCAAAAT................................................................................................ | 22 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAATAG.............................................................................................. | 23 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAATAGT............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAA................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAAa................................................................................................ | 21 | A | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAA................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAA.................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................AATGTAGAGATTGATCAAAAaa............................................................................................... | 22 | AA | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................AATGTAGAGATTGATCAAAATt............................................................................................... | 22 | T | 1.00 | 37.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............TATTTAAAAATGTCTgaac............................................................................................................................................. | 19 | GAAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................AATGTAGAGATTGATCAca.................................................................................................. | 19 | CA | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TAACTATTTTACAACTATTTAAAAATGTCTTAGAGTTTAACTGGCCTAAAATATATGAAATGTAGAGATTGATCAAAATAGTTTCTATCAAAATAGTTTTGATCAATCTCTGCAATTTTATATATGAGGAAACTGAAGTCTGAAAGTATTATGCCCTTTGTGTCTCTATTTTGAA ..................................................(((((((((((((((((((((((((((((.((((......)))).))))))))))))))))))).)))))))))).................................................. ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR343336 | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR040012(GSM532897) G648N. (cervix) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR343337 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................tATCAATCTCTGCAATTT......................................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................cgaAAACTGAAGTCTGAAAG............................. | 20 | cga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................ttAAACTGAAGTCTGAAAG............................. | 19 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |