| (1) AGO1.ip | (1) AGO2.ip | (7) BREAST | (23) CELL-LINE | (1) FIBROBLAST | (4) HELA | (4) KIDNEY | (5) LIVER | (1) LUNG | (1) OTHER | (2) OVARY | (2) SKIN |
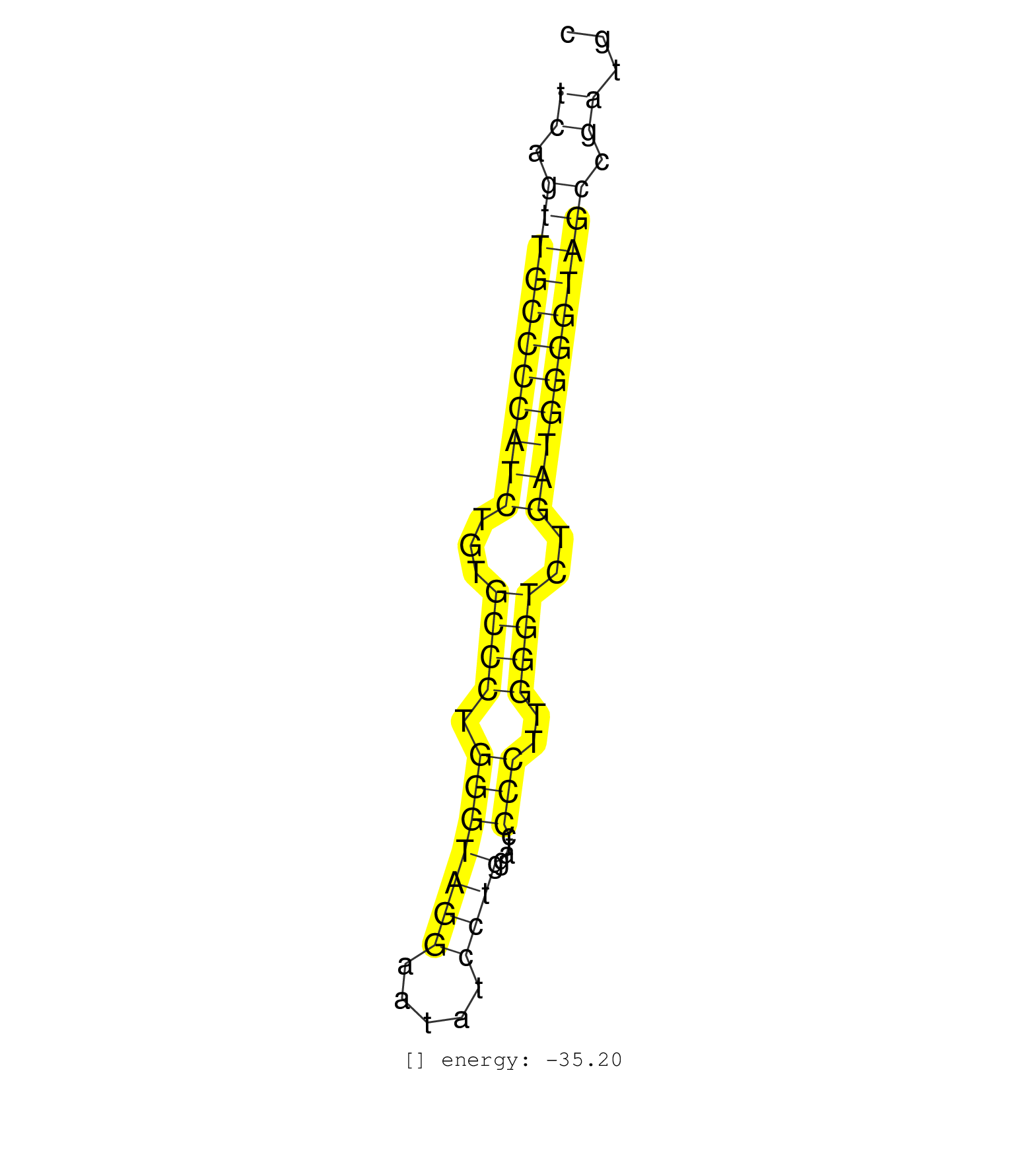
| CATATCTAATCAGGGATTCCTCATCTTGAAAAGCCCAGACCTACCTTTGAGCCTCAGTTGCCCCATCTGTGCCCTGGGTAGGAATATCCTGGATCCCCTTGGGTCTGATGGGGTAGCCGATGCCTGATTTGCACCCACAACGTGGGAGGTTATAACCTGTCCCCAAGTTGCAA ..................................................((.(((((((((((...((((.(((((((.....))))....)))..))))..))))))))))).))..................................................... .....................................................54...................................................................123................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR038863(GSM458546) MM603. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029130(GSM416759) DLD2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR191611(GSM715721) 57genomic small RNA (size selected RNA from t. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR038858(GSM458541) MEL202. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR139178(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | GSM339994(GSM339994) hues6. (cell line) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR038860(GSM458543) MM426. (cell line) | SRR191558(GSM715668) 61genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGC........................................................ | 22 | 1 | 60.00 | 60.00 | - | - | 16.00 | 5.00 | 8.00 | 5.00 | 1.00 | 8.00 | 2.00 | 2.00 | 2.00 | 4.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAG......................................................... | 21 | 1 | 58.00 | 58.00 | 8.00 | 12.00 | 6.00 | 6.00 | 3.00 | 4.00 | - | - | 4.00 | - | - | 1.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 |
| ..........................................................TGCCCCATCTGTGCCCTGGGTAGG........................................................................................... | 24 | 1 | 27.00 | 27.00 | 16.00 | - | - | - | 2.00 | 1.00 | 1.00 | 1.00 | - | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGt........................................................ | 22 | T | 15.00 | 58.00 | - | 2.00 | - | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTA.......................................................... | 20 | 1 | 10.00 | 10.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTAGGA.......................................................................................... | 25 | 1 | 7.00 | 7.00 | - | - | 1.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGa........................................................ | 22 | A | 6.00 | 58.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCat...................................................... | 24 | AT | 6.00 | 60.00 | - | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGGTCTGATGGGGTAGCCGAT.................................................... | 22 | 1 | 5.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTAG............................................................................................ | 23 | 1 | 5.00 | 5.00 | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCt....................................................... | 23 | T | 5.00 | 60.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCa....................................................... | 23 | A | 4.00 | 60.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGta....................................................... | 23 | TA | 4.00 | 58.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................TGGGTCTGATGGGGTAGCCGttt................................................... | 23 | TTT | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGaat...................................................... | 24 | AAT | 3.00 | 58.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGaaa...................................................... | 24 | AAA | 3.00 | 58.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCaa...................................................... | 24 | AA | 2.00 | 60.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGtt....................................................... | 23 | TT | 2.00 | 58.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTAGa........................................................................................... | 24 | A | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGaa....................................................... | 23 | AA | 2.00 | 58.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCtta..................................................... | 25 | TTA | 2.00 | 60.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCac...................................................... | 24 | AC | 1.00 | 60.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGCCGA..................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGcagc........................................................ | 22 | CAGC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGCatt..................................................... | 25 | ATT | 1.00 | 60.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CTTGGGTCTGATGGGGTAGC........................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CTTGAAAAGCCCAGAgatt.................................................................................................................................. | 19 | GATT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAac........................................................ | 22 | AC | 1.00 | 10.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGtttt..................................................... | 25 | TTTT | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGtat...................................................... | 24 | TAT | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGT........................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTgag........................................................................................... | 24 | GAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGCCGt..................................................... | 22 | T | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGCttt..................................................... | 22 | TTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCTTGGGTCTGATGGGGTAGC........................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTA.......................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGCCGAaa................................................... | 24 | AA | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGa........................................................ | 19 | A | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GCCCCATCTGTGCCCTGGGTAGG........................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAGCCGAT.................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGtag...................................................... | 24 | TAG | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TTGGGTCTGATGGGGTAG......................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ATCTTGAAAAGCCCAGAgata.................................................................................................................................. | 21 | GATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................TGGGTCTGATGGGGTA.......................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTAGGAA......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGGTCTGATGGGGTAGa........................................................ | 18 | A | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GCCCCATCTGTGCCCTGGGTAGGt.......................................................................................... | 24 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGaca...................................................... | 24 | ACA | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCCCATCTGTGCCCTGGGTga............................................................................................ | 23 | GA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGtta...................................................... | 24 | TTA | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGttat..................................................... | 25 | TTAT | 1.00 | 58.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGGGTAGag....................................................... | 23 | AG | 1.00 | 58.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGGGTCTGATGGGGTAGCCGAaa................................................... | 23 | AA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCCTTGGGTCTGATGGG............................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CATATCTAATCAGGGATTCCTCATCTTGAAAAGCCCAGACCTACCTTTGAGCCTCAGTTGCCCCATCTGTGCCCTGGGTAGGAATATCCTGGATCCCCTTGGGTCTGATGGGGTAGCCGATGCCTGATTTGCACCCACAACGTGGGAGGTTATAACCTGTCCCCAAGTTGCAA ..................................................((.(((((((((((...((((.(((((((.....))))....)))..))))..))))))))))).))..................................................... .....................................................54...................................................................123................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR038863(GSM458546) MM603. (cell line) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029130(GSM416759) DLD2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR191611(GSM715721) 57genomic small RNA (size selected RNA from t. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR038858(GSM458541) MEL202. (cell line) | SRR029127(GSM416756) A549. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR139178(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | GSM339994(GSM339994) hues6. (cell line) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR038860(GSM458543) MM426. (cell line) | SRR191558(GSM715668) 61genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................acccTGAGCCTCAGTTGCCC.............................................................................................................. | 20 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................actgCCTGGATCCCCTTGG....................................................................... | 19 | actg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................agccTATCCTGGATCCCCT.......................................................................... | 19 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |