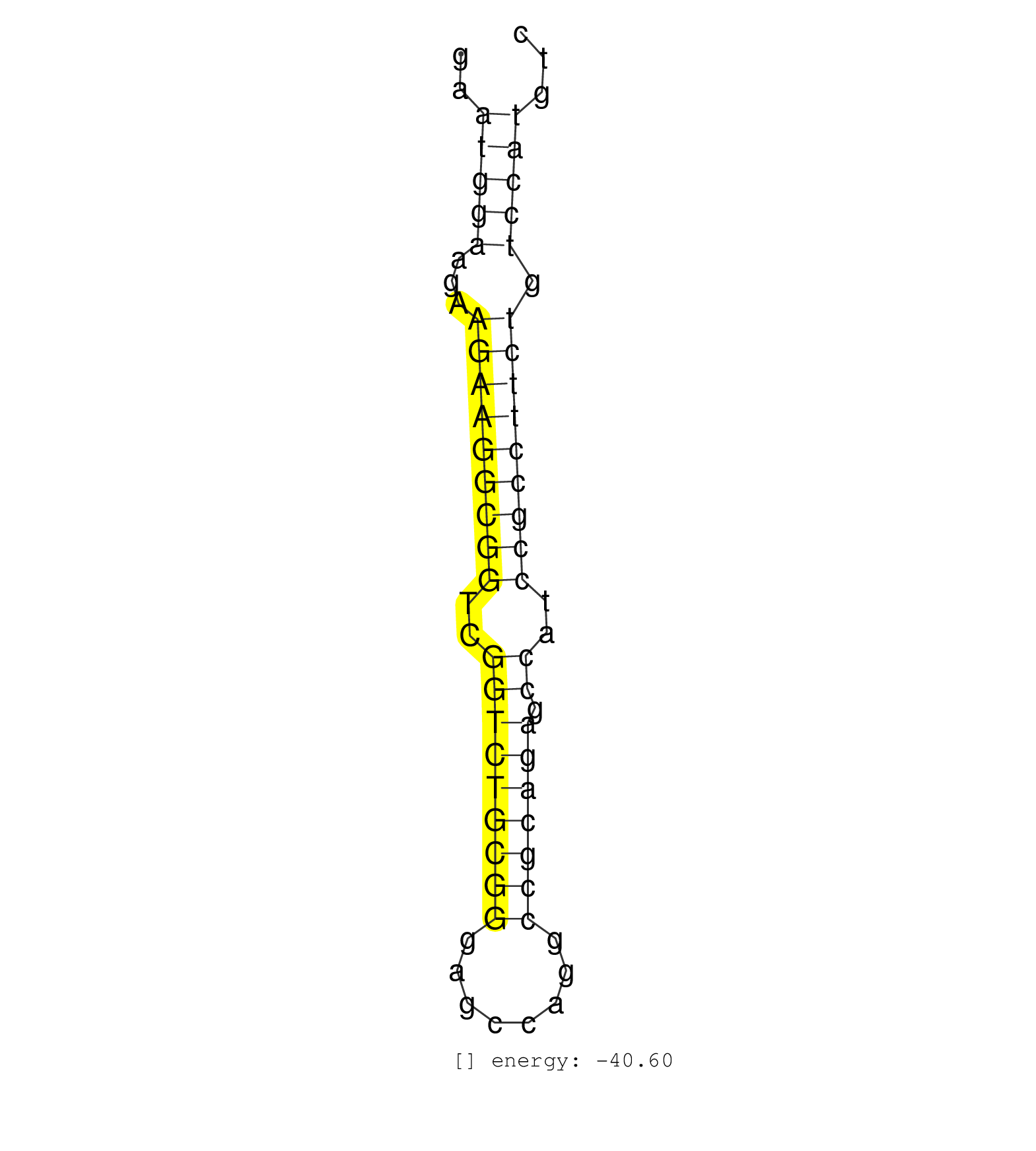
| (1) AGO2.ip | (1) B-CELL | (2) BREAST | (6) CELL-LINE | (1) HEART | (2) OTHER | (3) SKIN | (1) XRN.ip |
| CCCCCGAGAGACCCAGGAAGAAGGCAAGGAGGTTCGGTACGAGCCATCTCGAATGGAAGAAGAAGGCGGTCGGTCTGCGGGAGCCAGGCCGCAGAGCCATCCGCCTTCTGTCCATGTCTTCTTTATCTTCCTCGCCATAATAATTAGTGGTGTGGACCCCCCCACACA ....................................................(((((...(((((((((..(((((((((........))))))).))..))))))))).)))))..................................................... ..................................................51.................................................................118................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR038859(GSM458542) MM386. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR189786 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR189783 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................AAGAAGGCGGTCGGTCTGCGG........................................................................................ | 21 | 1 | 5.00 | 5.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCG......................................................................................... | 22 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCGG........................................................................................ | 23 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................AGAAGGCAAGGAGGTccgg................................................................................................................................... | 19 | CCGG | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCGGaa...................................................................................... | 25 | AA | 2.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GAAGAAGGCAAGGAGGTTCGGTACGAGCCATC........................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCGa........................................................................................ | 23 | A | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GCGGTCGGTCTGCGGcggt.................................................................................... | 19 | CGGT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGC.......................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCGtat...................................................................................... | 25 | TAT | 1.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................CATCTCGAATGGAAGAAGAAGGC..................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCGaat...................................................................................... | 25 | AAT | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................AGAAGAAGGCGGTCGGTCTGCc......................................................................................... | 22 | C | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AAGGCAAGGAGGTTCGGT.................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| .............................................ATCTCGAATGGAAGA............................................................................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .....................................................TGGAAGAAGAAGGCG.................................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |
| CCCCCGAGAGACCCAGGAAGAAGGCAAGGAGGTTCGGTACGAGCCATCTCGAATGGAAGAAGAAGGCGGTCGGTCTGCGGGAGCCAGGCCGCAGAGCCATCCGCCTTCTGTCCATGTCTTCTTTATCTTCCTCGCCATAATAATTAGTGGTGTGGACCCCCCCACACA ....................................................(((((...(((((((((..(((((((((........))))))).))..))))))))).)))))..................................................... ..................................................51.................................................................118................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR038859(GSM458542) MM386. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR189786 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR189783 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................gtgcCTGCGGGAGCCAGGCC.............................................................................. | 20 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................cTCCGCCTTCTGTCCA...................................................... | 16 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................CCGCAGAGCCATCCGCCTTCTGT......................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCTTCTTTATCTTCCTC................................... | 17 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |