| (2) AGO2.ip | (1) B-CELL | (6) BRAIN | (28) BREAST | (20) CELL-LINE | (3) CERVIX | (2) HEART | (1) LIVER | (1) OTHER | (1) RRP40.ip | (2) SKIN | (1) UTERUS |
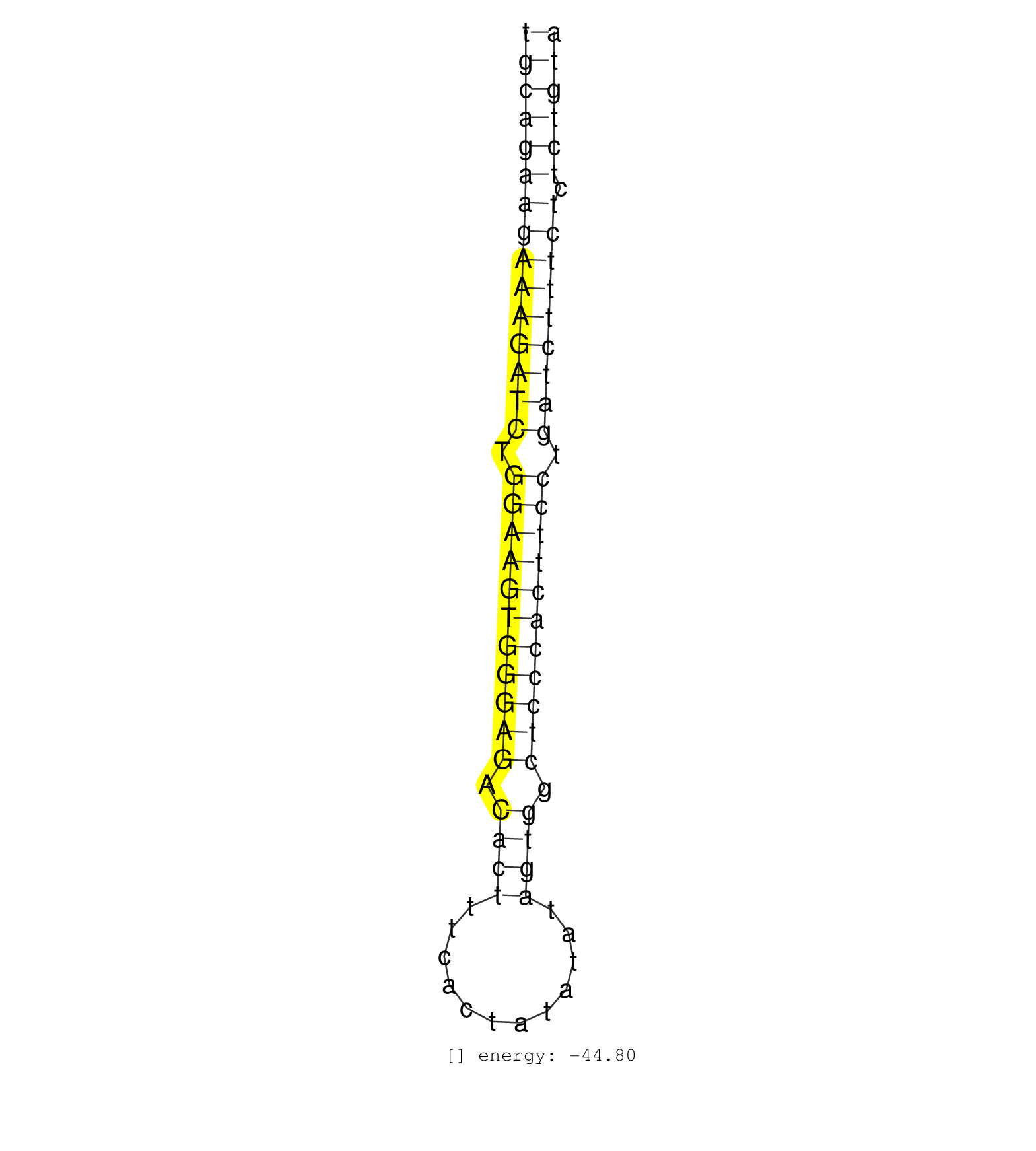
| TGTGCTTAGATGAGCTAAATGTTTAAAGGTTGGAGACTGCCATGAAGCTCTGCAGAAGAAAGATCTGGAAGTGGGAGACACTTTCACTATATATAGTGGCTCCCACTTCCTGATCTTTCTCTCTGTATATATAGTACTTAGAGAAATTCAACTATCAGGACTCAGTTTTTCTAGCAG ..................................................(((((((((((((((.(((((((((((.((((............)))).))))))))))).))))))))).)))))).................................................. ..................................................51..........................................................................127................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191401(GSM715511) 38genomic small RNA (size selected RNA from t. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR038863(GSM458546) MM603. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR029128(GSM416757) H520. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR038860(GSM458543) MM426. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191480(GSM715590) 38genomic small RNA (size selected RNA from t. (breast) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | TAX577589(Rovira) total RNA. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040042(GSM532927) G428N. (cervix) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR038855(GSM458538) D10. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191409(GSM715519) 19genomic small RNA (size selected RNA from t. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR191556(GSM715666) 45genomic small RNA (size selected RNA from t. (breast) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR191466(GSM715576) 114genomic small RNA (size selected RNA from . (breast) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | SRR040014(GSM532899) G623N. (cervix) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR191394(GSM715504) 23genomic small RNA (size selected RNA from t. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) | SRR191589(GSM715699) 69genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................AAAGATCTGGAAGTGGGAGAC.................................................................................................. | 21 | 3 | 18.33 | 18.33 | 4.33 | 2.33 | 1.00 | - | 0.33 | 0.33 | 0.67 | - | 1.67 | 0.67 | 1.67 | 0.33 | 0.67 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | 0.33 | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGA................................................................................................... | 20 | 3 | 15.67 | 15.67 | 3.33 | 1.33 | 3.67 | - | - | 0.67 | - | - | - | 0.67 | - | 1.00 | 0.33 | 0.67 | - | 0.67 | 0.33 | - | - | - | 0.33 | 0.33 | - | - | - | 0.67 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | 0.33 | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACA................................................................................................. | 22 | 3 | 4.67 | 4.67 | 1.00 | 0.67 | 0.33 | - | 1.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GAAGAAAGATCTGGAAGTGGG...................................................................................................... | 21 | 3 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACt................................................................................................. | 22 | T | 4.00 | 18.33 | 0.33 | 0.67 | 0.33 | - | - | 0.67 | 0.67 | - | 0.33 | - | - | - | - | - | 0.33 | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGTGGGA..................................................................................................... | 23 | 3 | 3.00 | 3.00 | - | 1.33 | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACc................................................................................................. | 22 | C | 2.33 | 18.33 | 1.00 | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGAaa................................................................................................. | 22 | AA | 1.33 | 15.67 | - | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGTG........................................................................................................ | 20 | 4 | 1.00 | 1.00 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACAa................................................................................................ | 23 | A | 1.00 | 4.67 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACAtc............................................................................................... | 24 | TC | 1.00 | 4.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGAa.................................................................................................. | 21 | A | 1.00 | 15.67 | 0.67 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGt................................................................................................... | 20 | T | 0.71 | 0.57 | 0.14 | 0.14 | - | - | - | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| ..........................................................AAAGATCTGGAAGTGGGAGAat................................................................................................. | 22 | AT | 0.67 | 15.67 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGCCATGAAGCTCTGCAGctc....................................................................................................................... | 21 | CTC | 0.67 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAAAGATCTGGAAGTGGGAGA................................................................................................... | 21 | 3 | 0.67 | 0.67 | - | - | - | - | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGAC.................................................................................................. | 20 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGAtt................................................................................................. | 22 | TT | 0.67 | 15.67 | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAG.................................................................................................... | 19 | 7 | 0.57 | 0.57 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| ........................................................................................................................CTCTGTATATATAGTAacc...................................... | 19 | ACC | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGACAt................................................................................................ | 22 | T | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACAC................................................................................................ | 23 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGATCTGGAAGTGGGAGACACT............................................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TCTGCAGAAGAAAGATCTGGAAGTGG....................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAAAGATCTGGAAGTGGGAGACc................................................................................................. | 23 | C | 0.33 | 0.00 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGA................................................................................................... | 19 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACAag............................................................................................... | 24 | AG | 0.33 | 4.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AAGAAAGATCTGGAAGTGGGAG.................................................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGTGG....................................................................................................... | 21 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGTGGGAGACA................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGAa.................................................................................................. | 20 | A | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTGGAAGTGGGAGAaca................................................................................................ | 18 | ACA | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AAGAAAGATCTGGAAGTGGGAGAa.................................................................................................. | 24 | A | 0.33 | 0.00 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGATCTGGAAGTGGGAGACAC................................................................................................ | 21 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGCCATGAAGCTCTGtcg.......................................................................................................................... | 18 | TCG | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGACt................................................................................................. | 21 | T | 0.33 | 0.67 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATCTGGAAGTGGGAGACACTT.............................................................................................. | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGAAAGATCTGGAAGTGGGAGA................................................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGTGGGAt.................................................................................................... | 24 | T | 0.33 | 3.00 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGATCTGGAAGTGGGAGACAa................................................................................................ | 21 | A | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGAt.................................................................................................. | 21 | T | 0.33 | 15.67 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAAGGTTGGAGACTGCtcct..................................................................................................................................... | 20 | TCCT | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAAAGGTTGGAGACTGCCATGAAGCTCTG............................................................................................................................. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGATCTGGAAGTGGGAGACA................................................................................................. | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACta................................................................................................ | 23 | TA | 0.33 | 18.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| .................................AGACTGCCATGAAGCaagc............................................................................................................................. | 19 | AAGC | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGACAC................................................................................................ | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAAAGATCTGGAAGTGGGAGAat................................................................................................. | 23 | AT | 0.33 | 0.67 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGAAGCTCTGCAGAAGcata................................................................................................................... | 20 | CATA | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGAaaaa............................................................................................... | 24 | AAAA | 0.33 | 15.67 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGCCATGAAGCTCTGtagg......................................................................................................................... | 19 | TAGG | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GAAGCTCTGCAGAAGAAgc................................................................................................................... | 19 | GC | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AAGATCTGGAAGTGGGAGACA................................................................................................. | 21 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATCTGGAAGTGGGAGACACT............................................................................................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GAAGAAAGATCTGGAAGTGGGAGAC.................................................................................................. | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGAAGAAAGATCTGGAAGT......................................................................................................... | 19 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| .............................................................GATCTGGAAGTGGGAagcc................................................................................................. | 19 | AGCC | 0.20 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAct................................................................................................... | 20 | CT | 0.20 | 0.00 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGgca................................................................................................. | 22 | GCA | 0.14 | 0.57 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGtca................................................................................................. | 22 | TCA | 0.14 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGgc.................................................................................................. | 21 | GC | 0.14 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGG...................................................................................................... | 17 | 9 | 0.11 | 0.11 | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGtgg................................................................................................... | 20 | TGG | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTGCTTAGATGAGCTAAATGTTTAAAGGTTGGAGACTGCCATGAAGCTCTGCAGAAGAAAGATCTGGAAGTGGGAGACACTTTCACTATATATAGTGGCTCCCACTTCCTGATCTTTCTCTCTGTATATATAGTACTTAGAGAAATTCAACTATCAGGACTCAGTTTTTCTAGCAG ..................................................(((((((((((((((.(((((((((((.((((............)))).))))))))))).))))))))).)))))).................................................. ..................................................51..........................................................................127................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191401(GSM715511) 38genomic small RNA (size selected RNA from t. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR038863(GSM458546) MM603. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR029128(GSM416757) H520. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR038860(GSM458543) MM426. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191480(GSM715590) 38genomic small RNA (size selected RNA from t. (breast) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | TAX577589(Rovira) total RNA. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040042(GSM532927) G428N. (cervix) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR038855(GSM458538) D10. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191409(GSM715519) 19genomic small RNA (size selected RNA from t. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | SRR191407(GSM715517) 81genomic small RNA (size selected RNA from t. (breast) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR191556(GSM715666) 45genomic small RNA (size selected RNA from t. (breast) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR191466(GSM715576) 114genomic small RNA (size selected RNA from . (breast) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | SRR040014(GSM532899) G623N. (cervix) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR191394(GSM715504) 23genomic small RNA (size selected RNA from t. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) | SRR191589(GSM715699) 69genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................aaagCTTCCTGATCTTTCT......................................................... | 19 | aaag | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGAAAGATCTGGAAGTGGGAG.................................................................................................... | 21 | 6 | 0.50 | 0.50 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAGACA................................................................................................. | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GAAAGATCTGGAAGTGGGAG.................................................................................................... | 20 | 6 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AAGAAAGATCTGGAAGTGGGAG.................................................................................................... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAGATCTGGAAGTGGGAG.................................................................................................... | 19 | 7 | 0.14 | 0.14 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |