| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) B-CELL | (4) BRAIN | (14) BREAST | (17) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (1) HEART | (1) KIDNEY | (5) LIVER | (1) LUNG | (3) OTHER | (2) OVARY | (1) RRP40.ip | (30) SKIN | (1) TESTES | (2) THYMUS | (3) UTERUS | (1) XRN.ip |
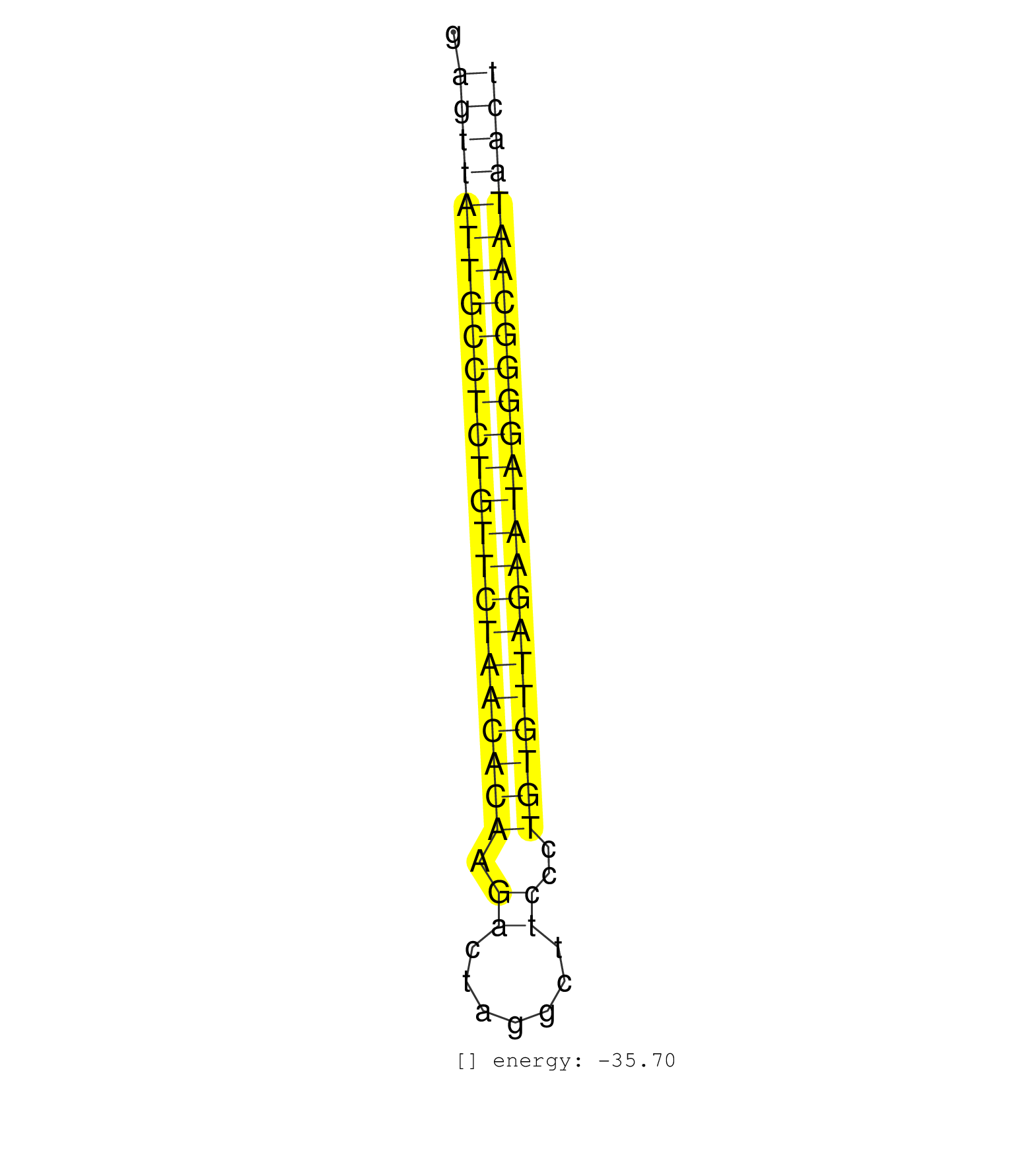
| ATATTTGGCCAGCATGAGTTCGTGAACTTCTCACAACAACCTTGAGAGCTGTGCAGAGTTATTGCCTCTGTTCTAACACAAGACTAGGCTTCCCTGTGTTAGAATAGGGGCAATAACTCTGCACTGCATTGCAGCCACCCACCCATCTAAGGAGGAAGTCCATGCTCTTTCCAT ...................................................((((((((((((((((((((((((.((.......))..)))))))))))))))))))))))).................................................. .......................................................56............................................................118...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR029128(GSM416757) H520. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189782 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040022(GSM532907) G575N. (cervix) | TAX577744(Rovira) total RNA. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191577(GSM715687) 93genomic small RNA (size selected RNA from t. (breast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | TAX577740(Rovira) total RNA. (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189785 | SRR191626(GSM715736) 36genomic small RNA (size selected RNA from t. (breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR191595(GSM715705) 72genomic small RNA (size selected RNA from t. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................ATTGCCTCTGTTCTAACACAAG............................................................................................ | 22 | 1 | 88.00 | 88.00 | 8.00 | 11.00 | 1.00 | 7.00 | - | 2.00 | 8.00 | 7.00 | - | 6.00 | - | - | 1.00 | - | - | 2.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | 1.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAAT............................................................ | 20 | 1 | 20.00 | 20.00 | 4.00 | - | 2.00 | - | 4.00 | - | - | - | 3.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATAA.......................................................... | 22 | 1 | 17.00 | 17.00 | - | - | 1.00 | - | - | 3.00 | - | - | - | - | 1.00 | 1.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATA........................................................... | 21 | 1 | 12.00 | 12.00 | 2.00 | - | - | - | - | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACAA............................................................................................. | 21 | 1 | 9.00 | 9.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................TTATTGCCTCTGTTCTAACACA.............................................................................................. | 22 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACA.............................................................................................. | 20 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TGTTAGAATAGGGGCAATAACT........................................................ | 22 | 1 | 8.00 | 8.00 | 2.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATAAt......................................................... | 23 | T | 6.00 | 17.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGCCTCTGTTCTAACACAAGA........................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACAC............................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATAt.......................................................... | 22 | T | 4.00 | 12.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGC............................................................... | 17 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACAAGt........................................................................................... | 23 | T | 3.00 | 88.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGCCTCTGTTCTAACACAAG............................................................................................ | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACAAGA........................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTTAGAATAGGGGCAATA........................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TTATTGCCTCTGTTCTAACA................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACAAc............................................................................................ | 22 | C | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................CTGTGTTAGAATAGGGGCAAT............................................................ | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAA............................................................. | 19 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACAaaat............................................................................................ | 22 | AAAT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AATAGGGGCAATAACTtta..................................................... | 19 | TTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACAtaa............................................................................................. | 21 | TAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGCCTCTGTTCTAACACg.............................................................................................. | 19 | G | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GTGTTAGAATAGGGGCAATAA.......................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TTATTGCCTCTGTTCTAACAC............................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTTAGAATAGGGGCAATAAC......................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATAAaa........................................................ | 24 | AA | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CTGTGTTAGAATAGGGGCA.............................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCAATAc.......................................................... | 22 | C | 1.00 | 12.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GCCTCTGTTCTAACACAAG............................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACcaa.............................................................................................. | 20 | CAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACcag............................................................................................ | 22 | CAG | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGCCTCTGTTCTAACACA.............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CTGTGTTAGAATAGGGGCAA............................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATTGCCTCTGTTCTAACAC............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATTGCCTCTGTTCTAACACt.............................................................................................. | 20 | T | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGTTAGAATAGGGGCA.............................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTTAGAATAGGGGCAATAAta........................................................ | 22 | TA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTGCCTCTGTTCTAACACAA............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GTGTTAGAATAGGGGCAATA........................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AACTCTGCACTGCATT............................................ | 16 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATATTTGGCCAGCATGAGTTCGTGAACTTCTCACAACAACCTTGAGAGCTGTGCAGAGTTATTGCCTCTGTTCTAACACAAGACTAGGCTTCCCTGTGTTAGAATAGGGGCAATAACTCTGCACTGCATTGCAGCCACCCACCCATCTAAGGAGGAAGTCCATGCTCTTTCCAT ...................................................((((((((((((((((((((((((.((.......))..)))))))))))))))))))))))).................................................. .......................................................56............................................................118...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR029128(GSM416757) H520. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189782 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040022(GSM532907) G575N. (cervix) | TAX577744(Rovira) total RNA. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189784 | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191557(GSM715667) 57genomic small RNA (size selected RNA from t. (breast) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191577(GSM715687) 93genomic small RNA (size selected RNA from t. (breast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | TAX577740(Rovira) total RNA. (breast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR189785 | SRR191626(GSM715736) 36genomic small RNA (size selected RNA from t. (breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR191595(GSM715705) 72genomic small RNA (size selected RNA from t. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................tccAGTCCATGCTCTTTCCA. | 20 | tcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....tcgtCCAGCATGAGTTCGT....................................................................................................................................................... | 19 | tcgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................aATTGCCTCTGTTCTAACACA.............................................................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |