| (1) AGO2.ip | (8) B-CELL | (1) BRAIN | (5) BREAST | (23) CELL-LINE | (1) CERVIX | (3) HEART | (5) LIVER | (2) OTHER | (1) OVARY | (1) RRP40.ip | (2) SKIN | (1) SPLEEN | (1) UTERUS | (1) XRN.ip |
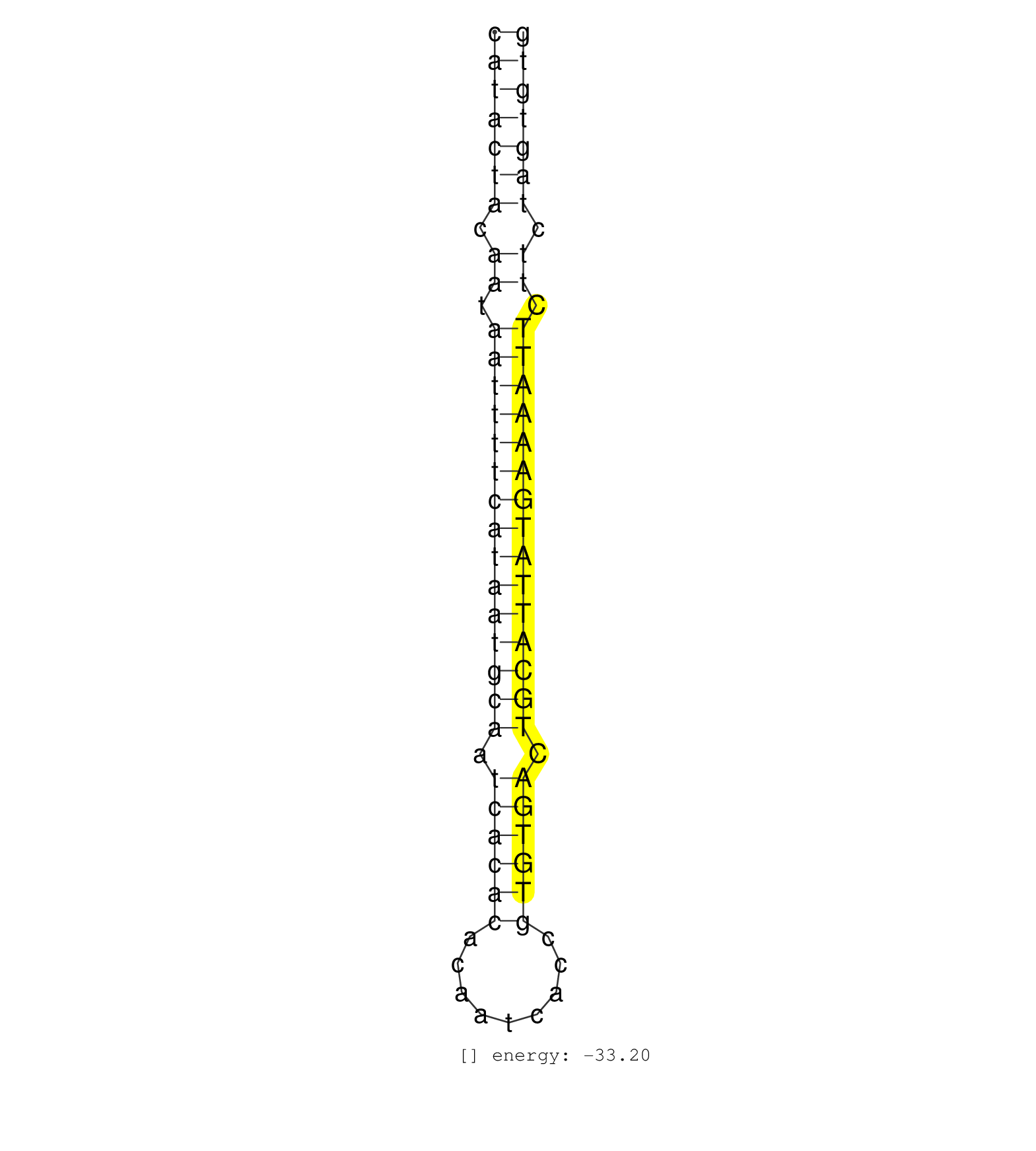
| GGACCCCAGCCTGGCCACATCTGCTTACAGGGCACTCTCAGGTGCCCACACATACTACAATAATTTTCATAATGCAATCACACACAATCACCGTGTGACTGCATTATGAAAATTCTTCTAGTGTGATTTACAGCTCTGTCAGGTCAGTTATTTTCTTCTTTATACTTGCTATTTT ..................................................(((((((.((.(((((((((((((((.((((((.........)))))).))))))))))))))).)).))))))).................................................. ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189782 | SRR029125(GSM416754) U2OS. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR038855(GSM458538) D10. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139164(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577739(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR038856(GSM458539) D11. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | TAX577579(Rovira) total RNA. (breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................TGTGACTGCATTATGAAAATTC............................................................ | 22 | 6 | 24.33 | 24.33 | 9.00 | 5.00 | 2.67 | 0.33 | 2.33 | - | 0.33 | - | - | 0.33 | 0.83 | 0.33 | 0.50 | 0.50 | 0.17 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.17 | 0.33 | - | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | 0.17 | 0.17 | 0.17 | - | - | 0.17 | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATT............................................................. | 21 | 6 | 10.00 | 10.00 | 4.17 | 0.17 | - | 1.00 | - | - | 1.00 | 1.33 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | 0.33 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | 0.17 | - | 0.17 | - | 0.17 | - | - | - | 0.17 | 0.17 | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAA............................................................... | 19 | 7 | 0.86 | 0.86 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.43 | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTt............................................................ | 22 | T | 0.67 | 10.00 | - | 0.17 | 0.33 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............GGCCACATCTGCTTACAGtgct............................................................................................................................................. | 22 | TGCT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAAT.............................................................. | 20 | 6 | 0.50 | 0.50 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACTGCATTATGAAAATTCTT.......................................................... | 22 | 6 | 0.50 | 0.50 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTCa........................................................... | 23 | A | 0.50 | 24.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATa............................................................. | 21 | A | 0.33 | 0.50 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTCg........................................................... | 23 | G | 0.33 | 24.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTTTCATAATGCAATCACACA........................................................................................... | 22 | 9 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTCc........................................................... | 23 | C | 0.17 | 24.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTa............................................................ | 22 | A | 0.17 | 10.00 | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GTGACTGCATTATGAAAATT............................................................. | 20 | 6 | 0.17 | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................ACTGCATTATGAAAATTattg......................................................... | 21 | ATTG | 0.17 | 0.00 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAATTta........................................................... | 23 | TA | 0.17 | 10.00 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAAgtc............................................................ | 22 | GTC | 0.14 | 0.86 | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGACCCCAGCCTGGCCACATCTGCTTACAGGGCACTCTCAGGTGCCCACACATACTACAATAATTTTCATAATGCAATCACACACAATCACCGTGTGACTGCATTATGAAAATTCTTCTAGTGTGATTTACAGCTCTGTCAGGTCAGTTATTTTCTTCTTTATACTTGCTATTTT ..................................................(((((((.((.(((((((((((((((.((((((.........)))))).))))))))))))))).)).))))))).................................................. ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189782 | SRR029125(GSM416754) U2OS. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR038862(GSM458545) MM472. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR038855(GSM458538) D10. (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139164(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577739(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR038856(GSM458539) D11. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | TAX577579(Rovira) total RNA. (breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TAATTTTCATAATGCAATCACA............................................................................................. | 22 | 7 | 2.43 | 2.43 | 0.43 | 0.14 | - | 0.29 | - | - | 0.14 | 0.14 | - | 0.71 | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.14 | 0.14 | - | - | - |
| .............................................................AATTTTCATAATGCAATCACA............................................................................................. | 21 | 10 | 1.20 | 1.20 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | 0.10 | - | - | - | - | - | - | - | 0.10 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | 0.10 | 0.10 |
| .....................................................................................ggggACCGTGTGACTGCATTAT.................................................................... | 22 | gggg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| caagCCCAGCCTGGCCACAT........................................................................................................................................................... | 20 | caag | 0.50 | 0.00 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CGTGTGACTGCATTATGAAAAT.............................................................. | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GCCACATCTGCTTACAGGGCACTCTCAGG..................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GTGACTGCATTATGAAAATTCT........................................................... | 22 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................tgacCAATCACACACAATCA..................................................................................... | 20 | tgac | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................aaACTCTCAGGTGCCCA............................................................................................................................... | 17 | aa | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GTGACTGCATTATGAAAAT.............................................................. | 19 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ttaTTTCATAATGCAATCACA............................................................................................. | 21 | tta | 0.20 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................agTAATTTTCATAATGCAATCA............................................................................................... | 22 | ag | 0.20 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................taaaTTTCATAATGCAATCACA............................................................................................. | 22 | taaa | 0.20 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................attaTTTCATAATGCAATCACA............................................................................................. | 22 | atta | 0.20 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGTGACTGCATTATGAAAAT.............................................................. | 20 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACTGCATTATGAAAATTCT........................................................... | 21 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................cTGTGACTGCATTATGAAAAT.............................................................. | 21 | c | 0.17 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................cacGTGACTGCATTATGAAAAT.............................................................. | 22 | cac | 0.17 | 0.33 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATTTTCATAATGCAATCACA............................................................................................. | 20 | 10 | 0.10 | 0.10 | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTTTCATAATGCAATCACA............................................................................................. | 19 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |