| (6) B-CELL | (1) BRAIN | (7) BREAST | (15) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (4) HEART | (1) KIDNEY | (1) LIVER | (3) OTHER | (13) SKIN |
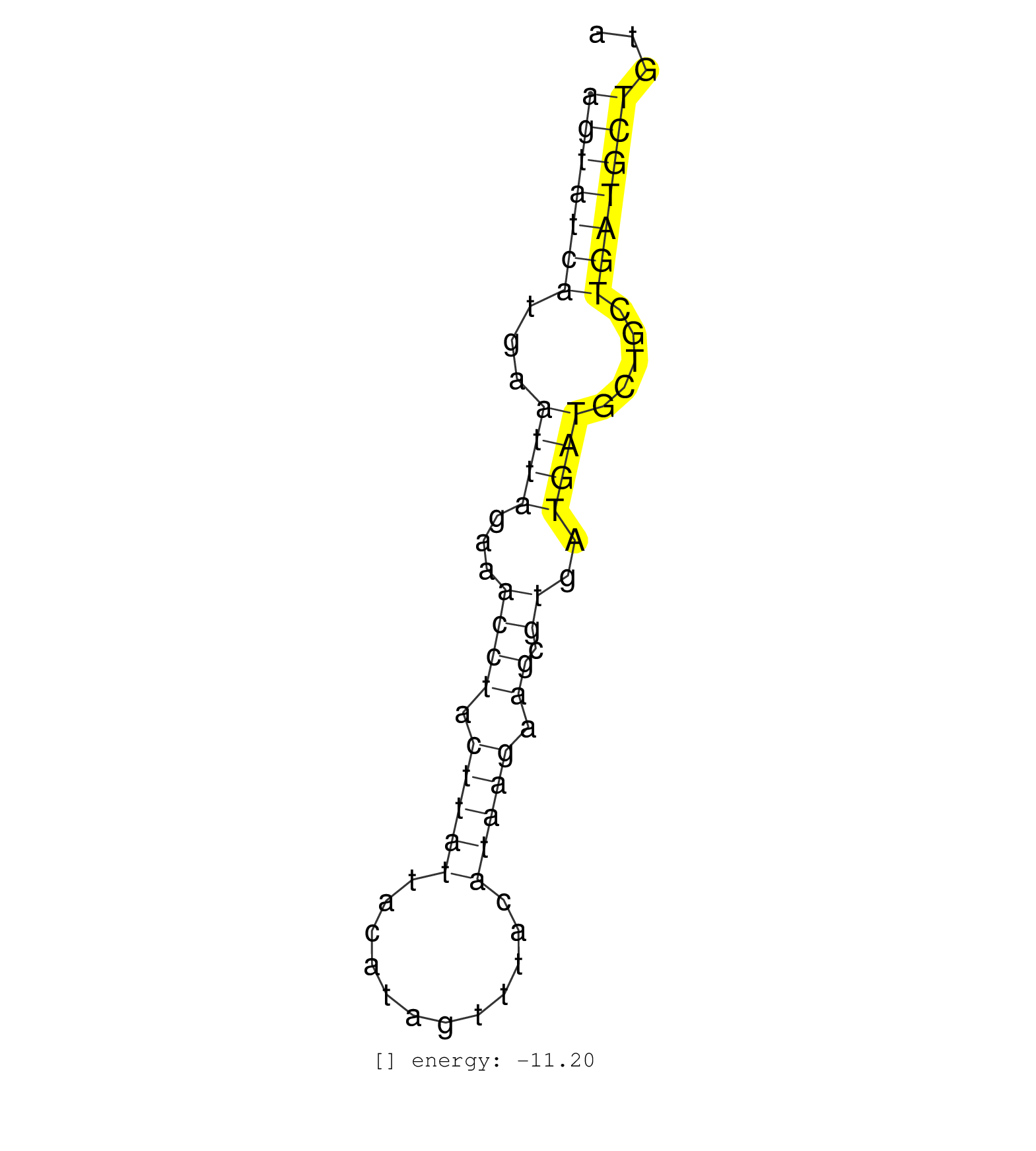
| AAGAAGCAATTTAACCTAATAGAACCAACTCCCCAGTTTGTCTGTAGAACAGTATCATGAATTAGAAACCTACTTATTACATAGTTTACATAAGAAGCGTGATGATGCTGCTGATGCTGTAATATCTAGTCTCTGTTGATGGTTCTTTCCTGGGAGGTTGGATGTGTTTCT ..................................................(((((((...((((...((((.(((((............))))).)).))..)))).....)))))))..................................................... ..................................................51....................................................................121................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR189785 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189784 | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR189782 | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM541796(GSM541796) undifferentiated human embryonic stem cells. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM532886(GSM532886) G850T. (cervix) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR191413(GSM715523) 28genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................ATGATGCTGCTGATGCTG.................................................... | 18 | 2 | 25.00 | 25.00 | 2.50 | 3.00 | - | 1.00 | 2.50 | 0.50 | - | - | 1.00 | 1.50 | 0.50 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | - | 0.50 | - | 0.50 | - | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | 0.50 | 0.50 | 0.50 | - | 0.50 | 0.50 | 0.50 | 0.50 | - | - | 0.50 | 0.50 | 0.50 |
| ...................................................................................................TGATGATGCTGCTGATGCTG.................................................... | 20 | 2 | 7.50 | 7.50 | - | - | - | - | 0.50 | 1.50 | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | 0.50 | - | 1.00 | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ....................................................................................................GATGATGCTGCTGATGCTG.................................................... | 19 | 2 | 5.00 | 5.00 | - | - | - | - | - | 0.50 | - | 2.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - |
| ..............................................................................................AAGCGTGATGATGCTGCTGATG....................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................GTATCATGAATTAGAt........................................................................................................ | 16 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................GTTTGTCTGTAGAACAGTATa................................................................................................................... | 21 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................GATGATGCTGCTGATGCTc.................................................... | 19 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGATGATGCTGCTGATGttt.................................................... | 20 | TTT | 0.50 | 0.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGAAGCAATTTAACCTAATAGAACCAACTCCCCAGTTTGTCTGTAGAACAGTATCATGAATTAGAAACCTACTTATTACATAGTTTACATAAGAAGCGTGATGATGCTGCTGATGCTGTAATATCTAGTCTCTGTTGATGGTTCTTTCCTGGGAGGTTGGATGTGTTTCT ..................................................(((((((...((((...((((.(((((............))))).)).))..)))).....)))))))..................................................... ..................................................51....................................................................121................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR189785 | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR189784 | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR189782 | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | GSM541796(GSM541796) undifferentiated human embryonic stem cells. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM532886(GSM532886) G850T. (cervix) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR191413(GSM715523) 28genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................gatcGTTTGTCTGTAGAAC......................................................................................................................... | 19 | gatc | 5.00 | 0.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................tcGTTTGTCTGTAGAAC......................................................................................................................... | 17 | tc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................cagcAACCAACTCCCCAGT...................................................................................................................................... | 19 | cagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................gggaAGTTTGTCTGTAGAA.......................................................................................................................... | 19 | ggga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................tatAGTTTGTCTGTAGAAC......................................................................................................................... | 19 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................gaacGTTTGTCTGTAGAAC......................................................................................................................... | 19 | gaac | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................gttcGTTTGTCTGTAGAAC......................................................................................................................... | 19 | gttc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ATGGTTCTTTCCTGGGA................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |