| (1) AGO2.ip | (15) B-CELL | (11) BRAIN | (16) BREAST | (24) CELL-LINE | (10) CERVIX | (1) FIBROBLAST | (5) HEART | (1) HELA | (10) LIVER | (2) OTHER | (1) OVARY | (1) PLACENTA | (1) RRP40.ip | (18) SKIN | (1) SPLEEN | (2) THYMUS | (4) UTERUS | (1) XRN.ip |
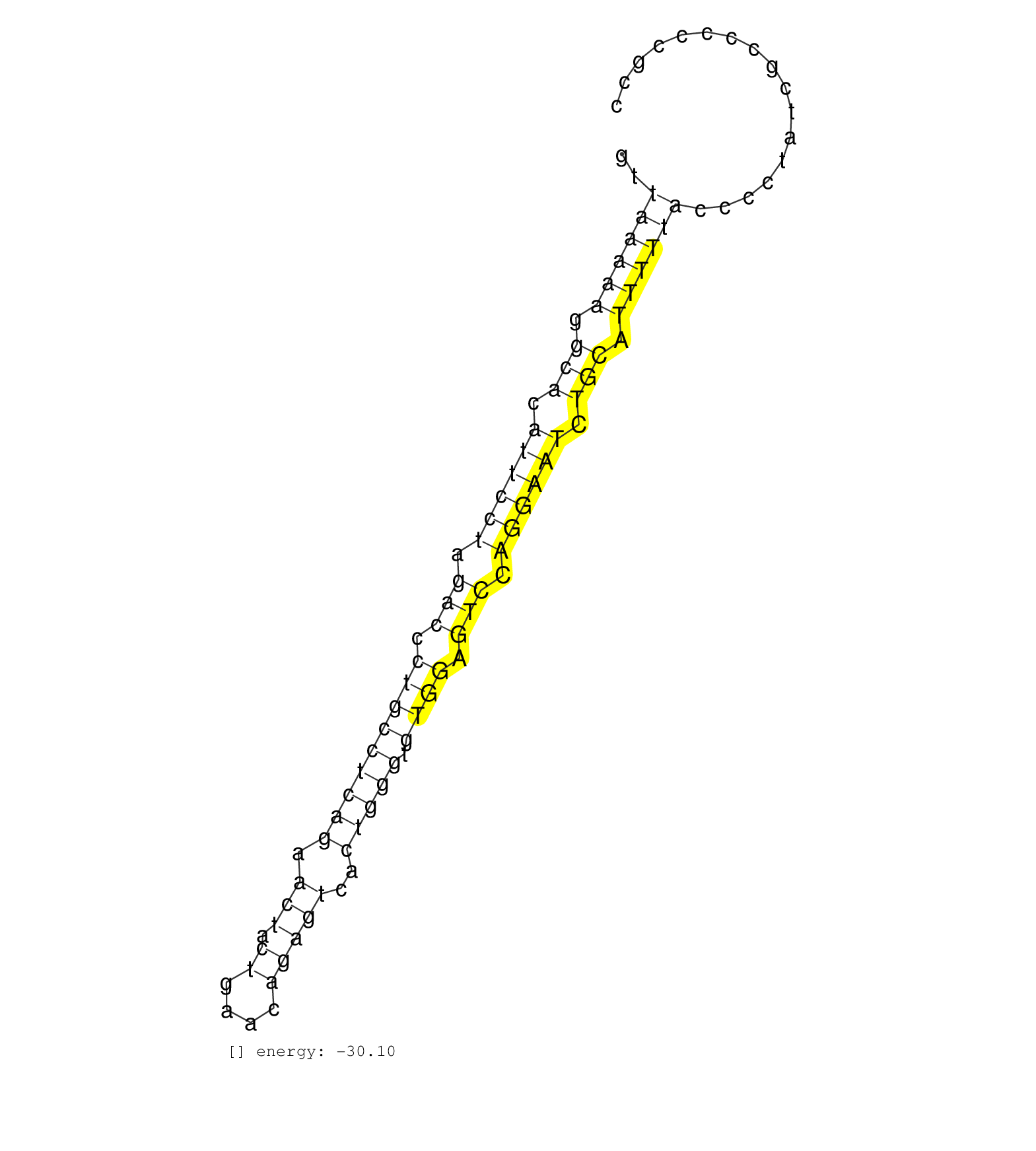
| AGGGCTTCTCAACATGTGATCTAAGACCACAGCTTTGTAACCTAACTCAGCCACGGTCCTAGTTAAAAAGGCACATTCCTAGACCCTGCCTCAGAACTACTGAACAGAGTCACTGGGTGTGGAGTCCAGGAATCTGCATTTTTACCCCTATCGCCCCCGCCATTCCCGCCCTTCCCCCCGCCATTGCGTGCACTAATTGCTGGAAGCACTC ....................................................((((((.(((.((((((.(((.(((((((((.(((.((....)))))..))))).)))).))).)))))).))).))))))................................................................... .............................................................62.................................................................................................161................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR040029(GSM532914) G026T. (cervix) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040018(GSM532903) G701N. (cervix) | SRR040008(GSM532893) G727N. (cervix) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | TAX577739(Rovira) total RNA. (breast) | GSM339994(GSM339994) hues6. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189784 | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR040030(GSM532915) G013N. (cervix) | GSM532877(GSM532877) G691N. (cervix) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR189787 | SRR040028(GSM532913) G026N. (cervix) | SRR040011(GSM532896) G529T. (cervix) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR189785 | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR040012(GSM532897) G648N. (cervix) | SRR040032(GSM532917) G603N. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029127(GSM416756) A549. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR139206(SRX050644) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR033728(GSM497073) MALT (MALT413). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR191599(GSM715709) 89genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR038854(GSM458537) MM653. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR191419(GSM715529) 41genomic small RNA (size selected RNA from t. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR191468(GSM715578) 116genomic small RNA (size selected RNA from . (breast) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR191481(GSM715591) 43genomic small RNA (size selected RNA from t. (breast) | SRR029128(GSM416757) H520. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR191617(GSM715727) 104genomic small RNA (size selected RNA from . (breast) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR038859(GSM458542) MM386. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR191562(GSM715672) 82genomic small RNA (size selected RNA from t. (breast) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTTT..................................................................... | 23 | 3 | 22.67 | 22.67 | - | 3.00 | 1.33 | - | 0.67 | - | - | - | - | 0.33 | 1.00 | 0.33 | - | 0.33 | 1.33 | 0.67 | - | - | 0.67 | 0.67 | 0.33 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | 0.33 | - | 0.67 | 0.67 | 0.67 | 0.67 | 0.67 | - | - | - | - | 0.33 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | - | 0.33 | - | 0.33 | 0.33 | 0.33 | 0.33 | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTT...................................................................... | 22 | 5 | 13.20 | 13.20 | - | - | 0.40 | - | 1.00 | - | - | - | - | 0.40 | - | 0.20 | 0.20 | - | - | 0.60 | 0.20 | 0.80 | 0.40 | 0.40 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.60 | 0.20 | 0.20 | - | - | - | - | - | - | 0.20 | 0.40 | - | 0.60 | - | 0.20 | 0.20 | - | - | - | - | 0.40 | 0.20 | 0.20 | 0.20 | 0.20 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | 0.20 | - | 0.20 | - | - | - | 0.20 | 0.20 | 0.20 | 0.20 | - | 0.20 | - | 0.20 | 0.20 | - | 0.20 | 0.20 | - | 0.20 | 0.20 | 0.20 | 0.20 | - | - | - | 0.20 | 0.20 | 0.20 | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCcggc............................ | 19 | CGGC | 8.00 | 0.00 | - | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TCAGAACTACTGAACtctg...................................................................................................... | 19 | TCTG | 6.00 | 0.00 | 2.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATT....................................................................... | 21 | 5 | 5.20 | 5.20 | - | - | 0.20 | - | 0.60 | - | - | - | - | - | 0.60 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | 0.20 | 0.20 | 0.60 | - | 0.20 | - | - | - | - | - | - | - | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | 0.20 | - | - | - | 0.20 | - | 0.20 | - | - | - | - | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GTGGAGTCCAGGAATCTGCATT....................................................................... | 22 | 3 | 3.33 | 3.33 | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 0.33 | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCcgg............................. | 18 | CGG | 3.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GAGTCCAGGAATCTGCATTTT..................................................................... | 21 | 4 | 2.00 | 2.00 | - | - | 1.25 | - | - | - | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTTTT.................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CCTCAGAACTACTGAACtctg...................................................................................................... | 21 | TCTG | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCcgcc............................ | 19 | CGCC | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCccg............................. | 18 | CCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CTCAGAACTACTGAACtctg...................................................................................................... | 20 | TCTG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GAGTCCAGGAATCTGCATTTTT.................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCctgc............................ | 19 | CTGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCAGGAATCTGCATTTTTcac................................................................. | 21 | CAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCccgg............................ | 19 | CCGG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................AGTCCAGGAATCTGCATT....................................................................... | 18 | 7 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCCGCCCTTCCCCCCGCCc............................ | 19 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTa...................................................................... | 22 | A | 0.80 | 5.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GGAGTCCAGGAATCTGCATTT...................................................................... | 21 | 5 | 0.60 | 0.60 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCAT........................................................................ | 20 | 7 | 0.57 | 0.57 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCcttt...................................................................... | 22 | CTTT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCA......................................................................... | 19 | 8 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.38 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - |
| ......................................................................................................................GTGGAGTCCAGGAATCTGCAT........................................................................ | 21 | 4 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GTGGAGTCCAGGAATCTGCATTT...................................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GTGGAGTCCAGGAATCTGCA......................................................................... | 20 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTca..................................................................... | 23 | CA | 0.20 | 5.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTTgt.................................................................... | 24 | GT | 0.20 | 13.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTag..................................................................... | 23 | AG | 0.20 | 5.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCATTc...................................................................... | 22 | C | 0.20 | 5.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................GAGTCCAGGAATCTGCATT....................................................................... | 19 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - |
| .......................................................................................................................TGGAGTCCAGGAATCTGCActt...................................................................... | 22 | CTT | 0.12 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| .....................................................................................................................................................................CCGCCCTTCCCCCCG............................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| AGGGCTTCTCAACATGTGATCTAAGACCACAGCTTTGTAACCTAACTCAGCCACGGTCCTAGTTAAAAAGGCACATTCCTAGACCCTGCCTCAGAACTACTGAACAGAGTCACTGGGTGTGGAGTCCAGGAATCTGCATTTTTACCCCTATCGCCCCCGCCATTCCCGCCCTTCCCCCCGCCATTGCGTGCACTAATTGCTGGAAGCACTC ....................................................((((((.(((.((((((.(((.(((((((((.(((.((....)))))..))))).)))).))).)))))).))).))))))................................................................... .............................................................62.................................................................................................161................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR040029(GSM532914) G026T. (cervix) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040018(GSM532903) G701N. (cervix) | SRR040008(GSM532893) G727N. (cervix) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | TAX577739(Rovira) total RNA. (breast) | GSM339994(GSM339994) hues6. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189784 | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR040030(GSM532915) G013N. (cervix) | GSM532877(GSM532877) G691N. (cervix) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR189787 | SRR040028(GSM532913) G026N. (cervix) | SRR040011(GSM532896) G529T. (cervix) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR189785 | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR040012(GSM532897) G648N. (cervix) | SRR040032(GSM532917) G603N. (cervix) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029127(GSM416756) A549. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR139206(SRX050644) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR033728(GSM497073) MALT (MALT413). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR191599(GSM715709) 89genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR038854(GSM458537) MM653. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR191419(GSM715529) 41genomic small RNA (size selected RNA from t. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR191468(GSM715578) 116genomic small RNA (size selected RNA from . (breast) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR191405(GSM715515) 55genomic small RNA (size selected RNA from t. (breast) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR191481(GSM715591) 43genomic small RNA (size selected RNA from t. (breast) | SRR029128(GSM416757) H520. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR191620(GSM715730) 167genomic small RNA (size selected RNA from . (breast) | SRR191617(GSM715727) 104genomic small RNA (size selected RNA from . (breast) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR038859(GSM458542) MM386. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR191562(GSM715672) 82genomic small RNA (size selected RNA from t. (breast) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR191600(GSM715710) 90genomic small RNA (size selected RNA from t. (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................ccgcTCCCGCCCTTCCCCC................................. | 19 | ccgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................ACCCTGCCTCAGAACTACTG............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................cgcTCCCGCCCTTCCCCCC................................ | 19 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................attCTAGACCCTGCCTCA...................................................................................................................... | 18 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................cctcCCCGCCCTTCCCCCC................................ | 19 | cctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................cAGTCCAGGAATCTGCAT........................................................................ | 18 | c | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................gtcGTCCAGGAATCTGCAT........................................................................ | 19 | gtc | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................ACATTCCTAGACCCTG........................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |