| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) AGO2.ip | (8) B-CELL | (5) BRAIN | (8) BREAST | (37) CELL-LINE | (3) CERVIX | (3) FIBROBLAST | (6) HEART | (1) HELA | (5) LIVER | (1) OTHER | (41) SKIN | (1) THYMUS | (1) XRN.ip |
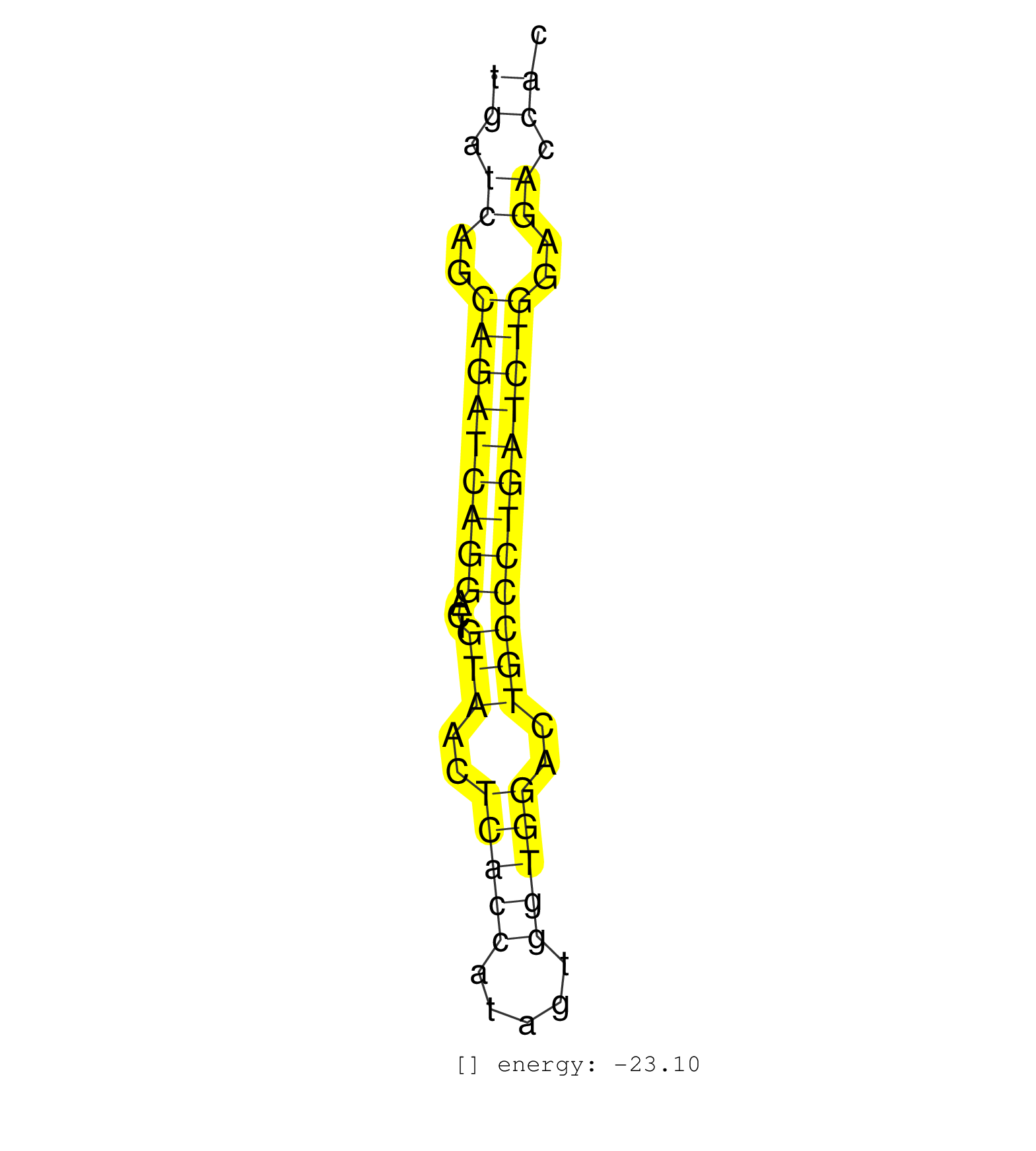
| CCCCCAGACAATGGAAGGGGGGCGACAGCTGTCCATGGGGGCACTGCTAAGAGGGTGTTGATCAGCAGATCAGGACTGTAACTCACCATAGTGGTGGACTGCCCTGATCTGGAGACCACTGCCTTTCGAGAATAATTCACCCTACAGAAGCTATCTGAGCATCACCAGACAGGGT ..................................................((.((..(((((((((...(((..(((((.....)))))..))))))))))))..)).))................................................... ..........................................................59..........................................................119...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038860(GSM458543) MM426. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR191427(GSM715537) 153genomic small RNA (size selected RNA from . (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040007(GSM532892) G601T. (cervix) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR040014(GSM532899) G623N. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038859(GSM458542) MM386. (cell line) | SRR330918(SRX091756) tissue: normal skindisease state: normal. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038853(GSM458536) MELB. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR038856(GSM458539) D11. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................TGGACTGCCCTGATCTGGAGA............................................................ | 21 | 1 | 187.00 | 187.00 | 44.00 | - | 7.00 | 7.00 | 3.00 | 4.00 | - | 4.00 | 4.00 | 6.00 | 4.00 | - | 3.00 | 1.00 | 1.00 | 3.00 | 4.00 | 4.00 | 4.00 | 2.00 | 3.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | 3.00 | 2.00 | 2.00 | 3.00 | 1.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | - | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAG............................................................. | 20 | 1 | 67.00 | 67.00 | 5.00 | - | 7.00 | 4.00 | 4.00 | 3.00 | - | 2.00 | 2.00 | - | - | 3.00 | - | 4.00 | 4.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 2.00 | - | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAGt............................................................ | 21 | T | 15.00 | 67.00 | 8.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTCACC........................................................................................ | 24 | 1 | 12.00 | 12.00 | 2.00 | 5.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTCA.......................................................................................... | 22 | 1 | 11.00 | 11.00 | - | 3.00 | - | - | - | - | 1.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTC........................................................................................... | 21 | 1 | 8.00 | 8.00 | - | 1.00 | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTCACCA....................................................................................... | 25 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAGAa........................................................... | 22 | A | 5.00 | 187.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTCAC......................................................................................... | 23 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAGAta.......................................................... | 23 | TA | 3.00 | 187.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................GTGGACTGCCCTGATCTGGAGA............................................................ | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAGAat.......................................................... | 23 | AT | 2.00 | 187.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................TGGACTGCCCTGATCTGGAGAaa.......................................................... | 23 | AA | 2.00 | 187.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....AGACAATGGAAGGGGtgat....................................................................................................................................................... | 19 | TGAT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAAC............................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTTCGAGAATAATTCACCCTACAGAA.......................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAta............................................................ | 21 | TA | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GTGGACTGCCCTGATCTGGAGAt........................................................... | 23 | T | 1.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AGGACTGTAACTCACCATAGTGGTGGAC............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGtagt............................................................ | 21 | TAGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGAGAtag......................................................... | 24 | TAG | 1.00 | 187.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GGACTGCCCTGATCTGGAG............................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AGCAGATCAGGACTGTAACTtact........................................................................................ | 24 | TACT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGtaga............................................................ | 21 | TAGA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGcg............................................................. | 20 | CG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGGA.............................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................GGGGGCACTGCTAAGggg......................................................................................................................... | 18 | GGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGcag............................................................. | 20 | CAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACTGCCCTGATCTGaaa............................................................. | 20 | AAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCACCCTACAGAAGCgcca.................... | 19 | GCCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCCCAGACAATGGAAGGGGGGCGACAGCTGTCCATGGGGGCACTGCTAAGAGGGTGTTGATCAGCAGATCAGGACTGTAACTCACCATAGTGGTGGACTGCCCTGATCTGGAGACCACTGCCTTTCGAGAATAATTCACCCTACAGAAGCTATCTGAGCATCACCAGACAGGGT ..................................................((.((..(((((((((...(((..(((((.....)))))..))))))))))))..)).))................................................... ..........................................................59..........................................................119...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038860(GSM458543) MM426. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR191427(GSM715537) 153genomic small RNA (size selected RNA from . (breast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR191485(GSM715595) 123genomic small RNA (size selected RNA from . (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040007(GSM532892) G601T. (cervix) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR040014(GSM532899) G623N. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038859(GSM458542) MM386. (cell line) | SRR330918(SRX091756) tissue: normal skindisease state: normal. (skin) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038853(GSM458536) MELB. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR038856(GSM458539) D11. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................ttcACTGCCCTGATCTGG............................................................... | 18 | ttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................cccGACTGCCCTGATCTG................................................................ | 18 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |