| (1) B-CELL | (5) CELL-LINE | (2) CERVIX | (1) HEART | (1) LIVER | (3) OTHER | (2) SKIN | (1) XRN.ip |
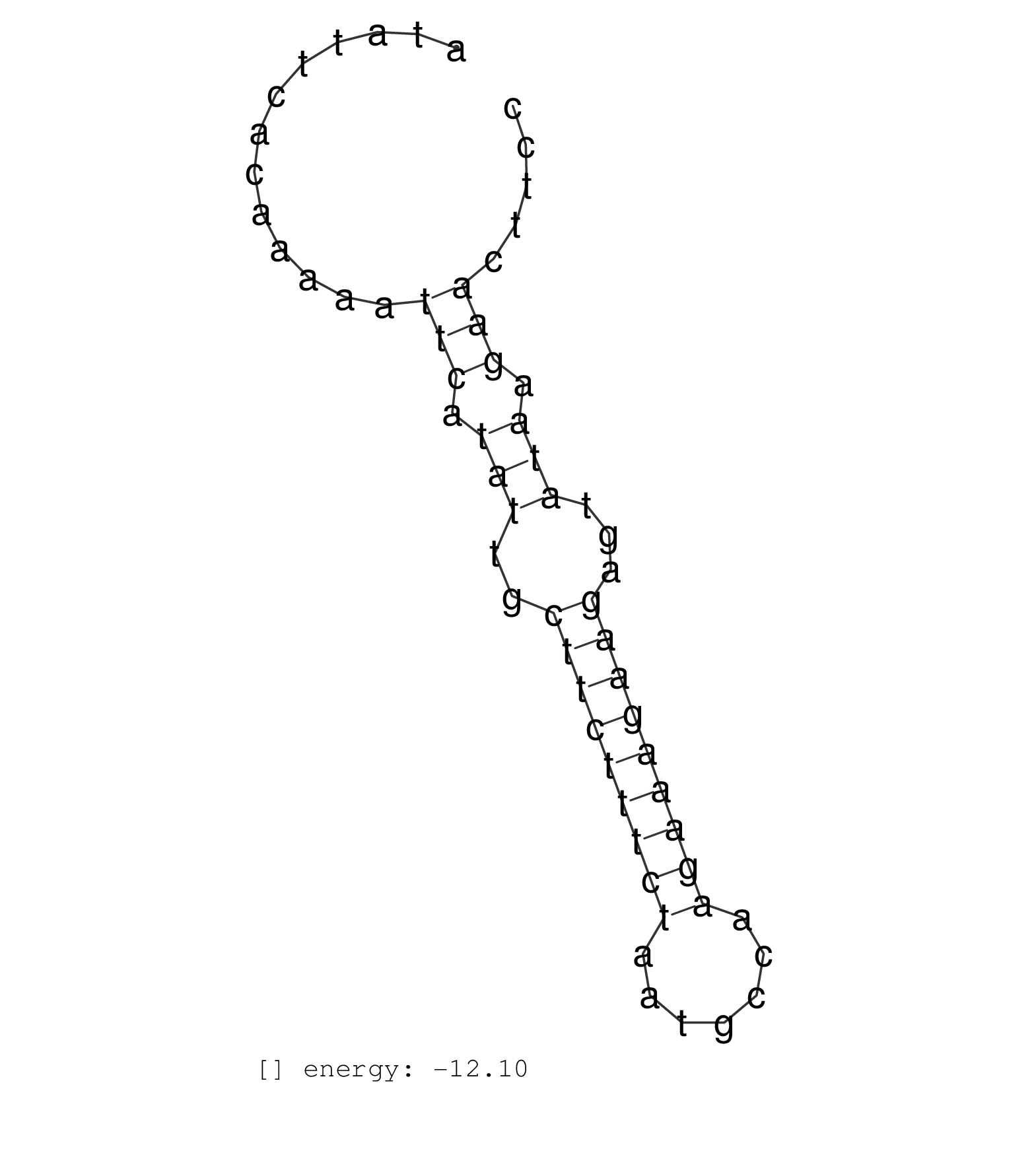
| GGTTAAGACAAACCTAAATCAATCTGAACAGTGTTGCCTGAATATTTAGAATATTCACAAAAATTCATATTGCTTCTTTCTAATGCCAAGAAAGAAGAGTATAAGAACTTCCTCCCAGAAGCCTAGACATTTTAATACATTTTCTATTTCAACTTACAATGA ...............................................................(((.(((..(((((((((.......)))))))))...))).)))....................................................... ..................................................51...........................................................112................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189785 | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR040039(GSM532924) G531T. (cervix) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................GAACAGTGTTGCCTGgtgt...................................................................................................................... | 19 | GTGT | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................TCCTCCCAGAAGCCTAtg................................... | 18 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................TCATATTGCTTCTTTCggt............................................................................... | 19 | GGT | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................ATTTTAATACATTTTCTttac............. | 21 | TTAC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AATATTTAGAATATTacga....................................................................................................... | 19 | ACGA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CTAAATCAATCTGAACtctg................................................................................................................................. | 20 | TCTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GGTTAAGACAAACCTAAATCAATCTGAACAGTGTTGCCTGAATATTTAGAATATTCACAAAAATTCATATTGCTTCTTTCTAATGCCAAGAAAGAAGAGTATAAGAACTTCCTCCCAGAAGCCTAGACATTTTAATACATTTTCTATTTCAACTTACAATGA ...............................................................(((.(((..(((((((((.......)))))))))...))).)))....................................................... ..................................................51...........................................................112................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189785 | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR040039(GSM532924) G531T. (cervix) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..ggttGACAAACCTAAATCAATCT......................................................................................................................................... | 23 | ggtt | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTTCCTCCCAGAAGCCTAGA................................... | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CAAGAAAGAAGAGTATAAGAA....................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................AATCTGAACAGTGTTGCCT........................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................ctCAAGAAAGAAGAGTATA........................................................... | 19 | ct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................ggACTTCCTCCCAGAAGCCTAGACATT............................... | 27 | gg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ctgcTTAAGACAAACCTAA................................................................................................................................................. | 19 | ctgc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TTTAGAATATTCACAAAAATT................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................cCTTCCTCCCAGAAGCCTAGACATTTT............................. | 27 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................gCTTCCTCCCAGAAGCCTAGACATTTT............................. | 27 | g | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CCTCCCAGAAGCCTAGAC.................................. | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| caTTAAGACAAACCTAAAT............................................................................................................................................... | 19 | ca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGAACAGTGTTGCCTGAAT....................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................ACTTCCTCCCAGAAGCCTAGACATTTT............................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........gCCTAAATCAATCTGAACAGTGTTGCCTGAA........................................................................................................................ | 31 | g | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................acaTGAATATTTAGAATAT............................................................................................................ | 19 | aca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................tCTTCCTCCCAGAAGCCTAGACATTTT............................. | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACTTCCTCCCAGAAGCCTAGACATTTTAAT.......................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................cCTTCCTCCCAGAAGCCTAGACATT............................... | 25 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CCTCCCAGAAGCCTAGACATTT.............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..ggttGACAAACCTAAATCAATCTGAACAGTGT................................................................................................................................ | 32 | ggtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................TGCCTGAATATTTAGA................................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .............CTAAATCAATCTGAA...................................................................................................................................... | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |