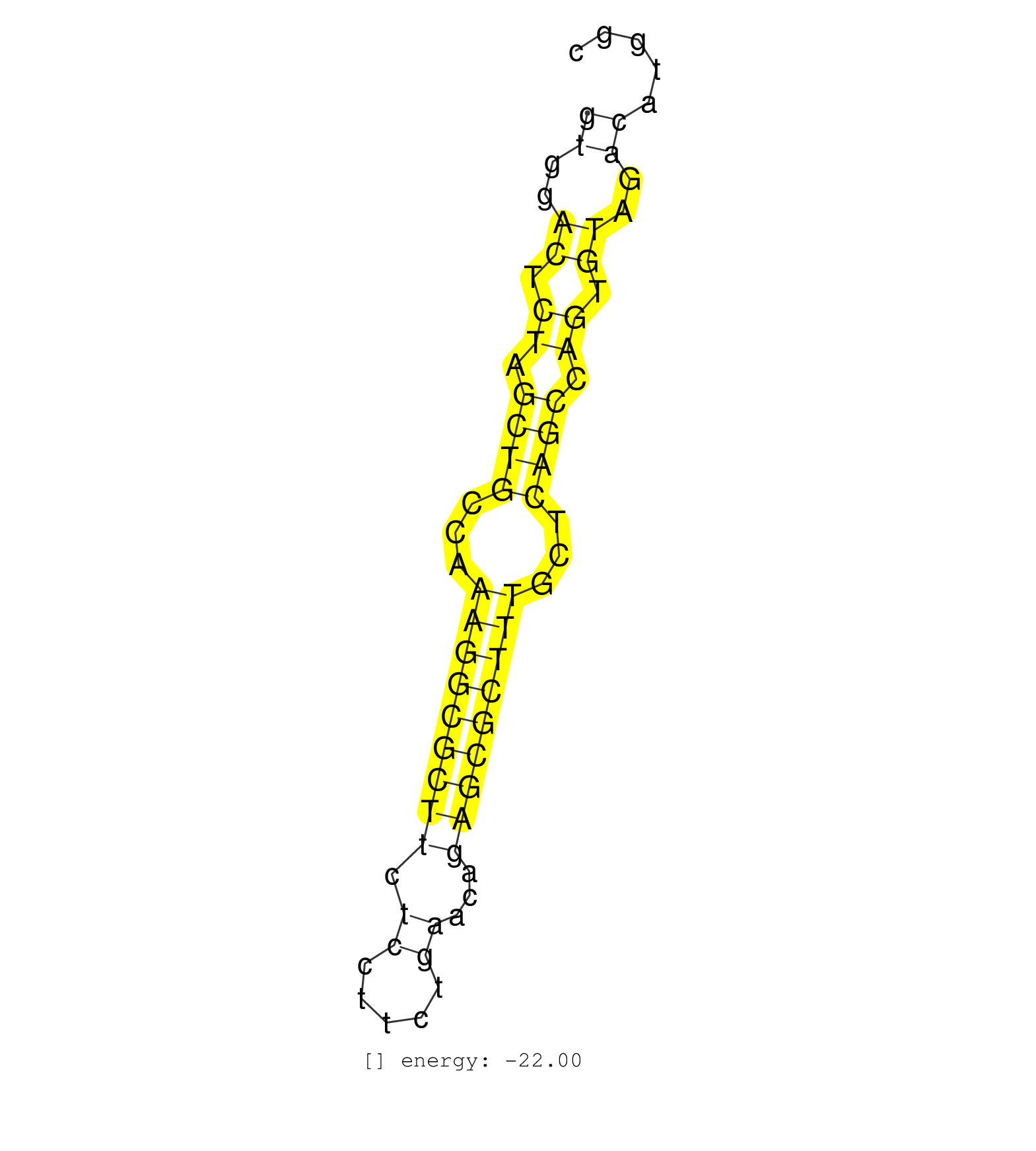
| (1) AGO2.ip | (1) B-CELL | (13) BRAIN | (29) BREAST | (11) CELL-LINE | (4) CERVIX | (3) FIBROBLAST | (8) HEART | (1) HELA | (5) KIDNEY | (1) LIVER | (4) LUNG | (2) OTHER | (2) OVARY | (3) PLACENTA | (3) SKIN | (4) TESTES | (1) UTERUS |
| CTGTGTAATTAGGTGAAGAAACTCCACCCGAGCGTCCATGGGTCAGCTATGTGGACTCTAGCTGCCAAAGGCGCTTCTCCTTCTGAACAGAGCGCTTTGCTCAGCCAGTGTAGACATGGCCTGATAAACAATGGAACTTCTCCCTTGAGAACTGGAGTTTGGATTGATCT ..................................................((..((.((.((((...(((((((((.((.....))...)))))))))...)))).)).))..))....................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR029131(GSM416760) MCF7. (cell line) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR139176(SRX050650) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR139183(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR029128(GSM416757) H520. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029125(GSM416754) U2OS. (cell line) | SRR191459(GSM715569) 32genomic small RNA (size selected RNA from t. (breast) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR191463(GSM715573) 111genomic small RNA (size selected RNA from . (breast) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139209(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040006(GSM532891) G601N. (cervix) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | TAX577743(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040040(GSM532925) G612N. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR191508(GSM715618) 152genomic small RNA (size selected RNA from . (breast) | SRR139185(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191581(GSM715691) 104genomic small RNA (size selected RNA from . (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR191588(GSM715698) 52genomic small RNA (size selected RNA from t. (breast) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040036(GSM532921) G243N. (cervix) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR191593(GSM715703) 62genomic small RNA (size selected RNA from t. (breast) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR189785 | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR191617(GSM715727) 104genomic small RNA (size selected RNA from . (breast) | SRR139186(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | SRR189786 | SRR139189(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR139162(SRX049422) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR191413(GSM715523) 28genomic small RNA (size selected RNA from t. (breast) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................ACTCTAGCTGCCAAAGGCGCT............................................................................................... | 21 | 1 | 826.00 | 826.00 | 568.00 | 41.00 | 32.00 | 21.00 | 18.00 | 11.00 | 15.00 | 9.00 | - | 7.00 | 9.00 | 10.00 | 7.00 | - | - | 7.00 | 6.00 | - | 1.00 | 5.00 | 5.00 | 6.00 | 4.00 | 1.00 | 4.00 | 5.00 | - | 3.00 | - | 1.00 | 1.00 | 3.00 | - | 1.00 | 1.00 | - | 2.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGC................................................................................................ | 20 | 1 | 419.00 | 419.00 | 375.00 | 9.00 | 4.00 | 2.00 | 2.00 | 2.00 | 3.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 3.00 | 2.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCG................................................................................................. | 19 | 1 | 207.00 | 207.00 | 175.00 | 5.00 | 5.00 | - | 5.00 | 4.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGC.................................................................................................. | 18 | 1 | 91.00 | 91.00 | 89.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTT.............................................................................................. | 22 | 1 | 44.00 | 44.00 | 7.00 | 3.00 | - | 6.00 | 3.00 | 1.00 | - | 6.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTt............................................................................................. | 23 | T | 15.00 | 44.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 3.00 | - | - | - | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTa............................................................................................. | 23 | A | 9.00 | 44.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGCGCTTTGCTCAGCCAGTGTAG......................................................... | 23 | 1 | 9.00 | 9.00 | 6.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CGCTTTGCTCAGCCAGTGTAGt........................................................ | 22 | T | 9.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTa.............................................................................................. | 22 | A | 9.00 | 826.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CGCTTTGCTCAGCCAGTGTAG......................................................... | 21 | 1 | 7.00 | 7.00 | - | - | 2.00 | - | - | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TCTAGCTGCCAAAGGCGCT............................................................................................... | 19 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGa................................................................................................ | 20 | A | 4.00 | 207.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCa............................................................................................... | 21 | A | 4.00 | 419.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCa................................................................................................. | 19 | A | 4.00 | 91.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGaa................................................................................................. | 19 | AA | 3.00 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 1.00 | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCactt.............................................................................................. | 22 | ACTT | 3.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTat............................................................................................. | 23 | AT | 3.00 | 826.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGat............................................................................................... | 21 | AT | 3.00 | 207.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGt................................................................................................ | 20 | T | 3.00 | 207.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTC............................................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAtgcg................................................................................................. | 19 | TGCG | 3.00 | 0.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTc.............................................................................................. | 22 | C | 3.00 | 826.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCg............................................................................................... | 21 | G | 3.00 | 419.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCTTTGCTCAGCCAGTGTAGt........................................................ | 23 | T | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGaaa................................................................................................. | 19 | AAA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTCTAGCTGCCAAAGGCGCT............................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGtcg................................................................................................. | 19 | TCG | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGaaa................................................................................................ | 20 | AAA | 2.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTCTa........................................................................................... | 25 | A | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTtt............................................................................................ | 24 | TT | 2.00 | 44.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCat.............................................................................................. | 22 | AT | 2.00 | 419.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GTCCATGGGTCAGCTATGTGG.................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGtcgc................................................................................................ | 20 | TCGC | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCtct............................................................................................... | 21 | TCT | 2.00 | 91.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCTCCTTCTGAACAGAG.............................................................................. | 17 | 3 | 1.67 | 1.67 | - | - | - | - | - | 0.33 | 1.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAagcg................................................................................................. | 19 | AGCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGtata................................................................................................ | 20 | TATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGacgc................................................................................................ | 20 | ACGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGCGCTTTGCTCAGCCAGgtag.......................................................... | 22 | GTAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCaa.............................................................................................. | 22 | AA | 1.00 | 419.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGCGCTTTGCTCAGCCAGTGTAGA........................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAtgct................................................................................................. | 19 | TGCT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCTTTGCTCAGCCAGTacg.......................................................... | 21 | ACG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGac.................................................................................................. | 18 | AC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TCTGAACAGAGCGCTTTGCTCAGCCAGTG............................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CGCTTTGCTCAGCCAGTGTAGtaa...................................................... | 24 | TAA | 1.00 | 7.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTCTAGCTGCCAAAGGCGCTTCT............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCgg.............................................................................................. | 22 | GG | 1.00 | 419.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCTTTGCTCAGCCAGTGTAGAa....................................................... | 24 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CCCTTGAGAACTGGAGTTTGG........ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCaaa............................................................................................... | 21 | AAA | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGccga................................................................................................ | 20 | CCGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTta............................................................................................ | 24 | TA | 1.00 | 44.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGCGCTTTGCTCAGCCAGTcat.......................................................... | 22 | CAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCact............................................................................................... | 21 | ACT | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CACCCGAGCGTCCATagc................................................................................................................................ | 18 | AGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CGCTTTGCTCAGCCAGTGTAGA........................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TCTAGCTGCCAAAGGCG................................................................................................. | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCTTTGCTCAGCCAGgtag.......................................................... | 21 | GTAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GCGCTTTGCTCAGCCAGTGTAGtt....................................................... | 24 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GAGCGCTTTGCTCAGCCAGTata.......................................................... | 23 | ATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAagca................................................................................................. | 19 | AGCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTg.............................................................................................. | 22 | G | 1.00 | 826.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTCT............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AAAGGCGCTTCTCCTTag...................................................................................... | 18 | AG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTTaa............................................................................................ | 24 | AA | 1.00 | 44.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGacgt................................................................................................ | 20 | ACGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCc............................................................................................... | 21 | C | 1.00 | 419.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................ATGTGGACTCTAGCTctg........................................................................................................ | 18 | CTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CAGCCAGTGTAGACAgct................................................... | 18 | GCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCctct............................................................................................ | 24 | CTCT | 1.00 | 419.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTctc............................................................................................ | 24 | CTC | 1.00 | 826.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCGCTgt............................................................................................. | 23 | GT | 1.00 | 826.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGacga................................................................................................ | 20 | ACGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGCtctt.............................................................................................. | 22 | TCTT | 1.00 | 91.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GAGCGCTTTGCTCAGCCAGTGTAG......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GCGCTTCTCCTTCTGAcggt................................................................................ | 20 | CGGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGtact............................................................................................... | 21 | TACT | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGagct............................................................................................... | 21 | AGCT | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGatag............................................................................................... | 21 | ATAG | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................ACTCTAGCTGCCAAAGGgg................................................................................................. | 19 | GG | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGGtgct............................................................................................... | 21 | TGCT | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ACTCTAGCTGCCAAAGG................................................................................................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCAGCCAGTGTAGACAT..................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| CTGTGTAATTAGGTGAAGAAACTCCACCCGAGCGTCCATGGGTCAGCTATGTGGACTCTAGCTGCCAAAGGCGCTTCTCCTTCTGAACAGAGCGCTTTGCTCAGCCAGTGTAGACATGGCCTGATAAACAATGGAACTTCTCCCTTGAGAACTGGAGTTTGGATTGATCT ..................................................((..((.((.((((...(((((((((.((.....))...)))))))))...)))).)).))..))....................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR029131(GSM416760) MCF7. (cell line) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139173(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR139176(SRX050650) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR139183(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | SRR040011(GSM532896) G529T. (cervix) | SRR029128(GSM416757) H520. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029125(GSM416754) U2OS. (cell line) | SRR191459(GSM715569) 32genomic small RNA (size selected RNA from t. (breast) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR191463(GSM715573) 111genomic small RNA (size selected RNA from . (breast) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139209(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040006(GSM532891) G601N. (cervix) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | TAX577743(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139168(SRX050648) FLASHPage purified small RNA (~15-40nt) from . (brain) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040040(GSM532925) G612N. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR191508(GSM715618) 152genomic small RNA (size selected RNA from . (breast) | SRR139185(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191581(GSM715691) 104genomic small RNA (size selected RNA from . (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR191588(GSM715698) 52genomic small RNA (size selected RNA from t. (breast) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040036(GSM532921) G243N. (cervix) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR191593(GSM715703) 62genomic small RNA (size selected RNA from t. (breast) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR189785 | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR191617(GSM715727) 104genomic small RNA (size selected RNA from . (breast) | SRR139186(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | SRR189786 | SRR139189(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR139162(SRX049422) FLASHPage purified small RNA (~15-40nt) from . (brain) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR191413(GSM715523) 28genomic small RNA (size selected RNA from t. (breast) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................ctctACTCCACCCGAGCGT....................................................................................................................................... | 19 | ctct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AACTTCTCCCTTGAG..................... | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |