| (3) B-CELL | (14) BRAIN | (14) BREAST | (4) CELL-LINE | (1) CERVIX | (3) FIBROBLAST | (3) HEART | (1) HELA | (7) LIVER | (1) OTHER | (1) OVARY | (2) PLACENTA | (37) SKIN | (2) TESTES | (4) UTERUS |
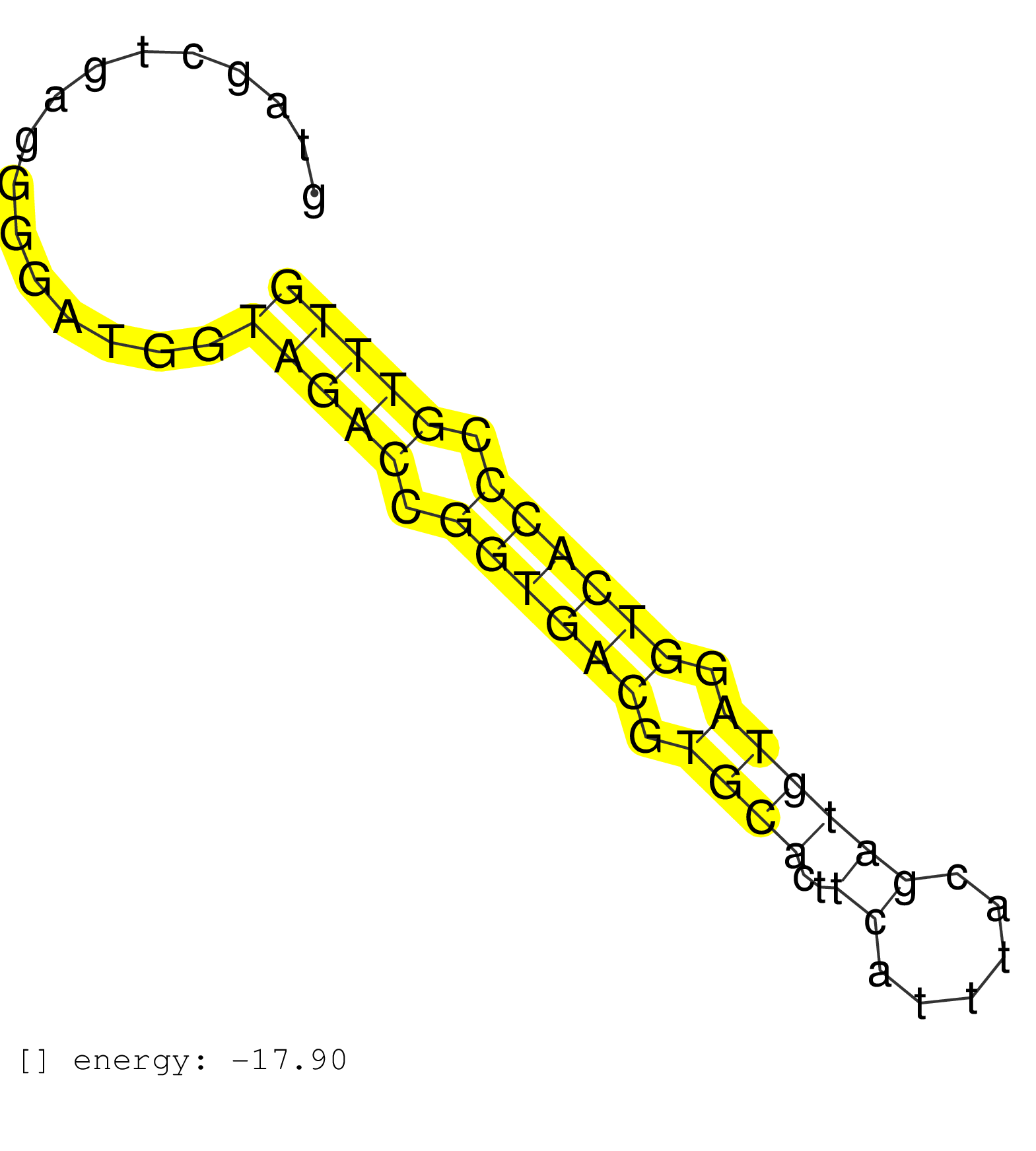
| ACACGCTAGCAACGTCATTGAAGGGACGATGGTGCCATCTGCTCTCCAGGGTAGCTGAGGGGATGGTAGACCGGTGACGTGCACTTCATTTACGATGTAGGTCACCCGTTTGACTATCCACCAGCGCCCGGTCCATCTGTGGGATGACATCTTGGTGCACATGGCAACTTCGGAGAAG ..................................................................(((((.((((((.((((..((......)))))).)))))).))))).................................................. ..................................................51...........................................................112................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR038854(GSM458537) MM653. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577741(Rovira) total RNA. (breast) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577579(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR040029(GSM532914) G026T. (cervix) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191487(GSM715597) 126genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR191623(GSM715733) 18genomic small RNA (size selected RNA from t. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................GGGATGGTAGACCGGTGACGTGC................................................................................................ | 23 | 1 | 196.00 | 196.00 | 41.00 | 36.00 | 16.00 | 10.00 | 10.00 | 1.00 | 2.00 | 9.00 | 3.00 | 4.00 | 4.00 | 5.00 | 5.00 | 3.00 | 2.00 | 4.00 | 2.00 | 3.00 | 1.00 | - | - | 1.00 | 1.00 | 4.00 | 1.00 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | 2.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCA............................................................................................... | 24 | 1 | 53.00 | 53.00 | 10.00 | 12.00 | 4.00 | 2.00 | 1.00 | 7.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | - | - | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAt.............................................................................................. | 25 | T | 20.00 | 53.00 | 5.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGt................................................................................................ | 23 | T | 18.00 | 6.00 | 1.00 | 3.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGT.................................................................................................. | 21 | 1 | 17.00 | 17.00 | 1.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCt............................................................................................... | 24 | T | 9.00 | 196.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATGGTAGACCGGTGACGTGC................................................................................................ | 21 | 1 | 8.00 | 8.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGa................................................................................................ | 23 | A | 8.00 | 6.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTG................................................................................................. | 22 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCA............................................................................................... | 20 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATGGTAGACCGGTGACGTGCA............................................................................................... | 21 | 1 | 5.00 | 5.00 | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGAC.................................................................................................... | 19 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCACTTC........................................................................................... | 24 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGggc................................................................................................ | 23 | GGC | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATGGTAGACCGGTGACGTGC................................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTaa................................................................................................ | 23 | AA | 3.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGtt............................................................................................... | 24 | TT | 3.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCGGTCCATCTGTGG.................................... | 15 | 2 | 2.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | 0.50 | 0.50 | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTat................................................................................................ | 23 | AT | 2.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CCATCTGTGGGATGACtga........................... | 19 | TGA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCAGCGCCCGGTCCAT.......................................... | 16 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTtt................................................................................................ | 23 | TT | 2.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGATGGTAGACCGGTGACGTGt................................................................................................ | 22 | T | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAta............................................................................................. | 26 | TA | 2.00 | 53.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAtt............................................................................................. | 26 | TT | 2.00 | 53.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACG................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATGGTAGACCGGTGACGTGCAt.............................................................................................. | 22 | T | 2.00 | 5.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTAT............................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATGGTAGACCGGTGACGTGCA............................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGcgc................................................................................................ | 23 | CGC | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCca.............................................................................................. | 25 | CA | 1.00 | 196.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACag.............................................................. | 19 | AG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAaat............................................................................................ | 27 | AAT | 1.00 | 53.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGagc................................................................................................ | 23 | AGC | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGATGGTAGACCGGTGACGTGCt............................................................................................... | 23 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CCATCTGTGGGATGAgc............................. | 17 | GC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAatt............................................................................................ | 27 | ATT | 1.00 | 53.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGgaa.............................................................................................. | 25 | GAA | 1.00 | 6.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTATCa........................................................... | 22 | A | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GGGGATGGTAGACCGGTGACGTGC................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGCAgat............................................................................................ | 27 | GAT | 1.00 | 53.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCAag............................................................................................. | 22 | AG | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTgca............................................................ | 21 | GCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCAa.............................................................................................. | 21 | A | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGATGGTAGACCGGTGACGTGCA............................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTATC............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATGGTAGACCGGTGACGTGCAa.............................................................................................. | 23 | A | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACatcc............................................................ | 21 | ATCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGATGGTAGACCGGTGACGTGC................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGggt................................................................................................ | 23 | GGT | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTtc................................................................................................ | 23 | TC | 1.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGta............................................................................................... | 24 | TA | 1.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATGGTAGACCGGTGACctgc................................................................................................ | 21 | CTGC | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATGGTAGACCGGTGACGTGCAa.............................................................................................. | 22 | A | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACctgc................................................................................................ | 23 | CTGC | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ACGATGGTGCCATCTGCTCTCCAGGGTc............................................................................................................................. | 28 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCACTTCt.......................................................................................... | 25 | T | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTATCC........................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGAa.................................................................................................... | 19 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGggca............................................................................................... | 24 | GGCA | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CCATCTGTGGGATGAtgac........................... | 19 | TGAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGGTCACCCGTTTGACTATCCA.......................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTGaa............................................................................................... | 24 | AA | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGTCACCCGTTTGACTATCt........................................................... | 22 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGATGGTAGACCGGTGACGTt................................................................................................. | 22 | T | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGTAGACCGGTGACGTGCACTT............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ATCTGTGGGATGACA............................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| ACACGCTAGCAACGTCATTGAAGGGACGATGGTGCCATCTGCTCTCCAGGGTAGCTGAGGGGATGGTAGACCGGTGACGTGCACTTCATTTACGATGTAGGTCACCCGTTTGACTATCCACCAGCGCCCGGTCCATCTGTGGGATGACATCTTGGTGCACATGGCAACTTCGGAGAAG ..................................................................(((((.((((((.((((..((......)))))).)))))).))))).................................................. ..................................................51...........................................................112................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR038854(GSM458537) MM653. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577741(Rovira) total RNA. (breast) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577579(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR040029(GSM532914) G026T. (cervix) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR191487(GSM715597) 126genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR191623(GSM715733) 18genomic small RNA (size selected RNA from t. (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................AGCGCCCGGTCCATCTGTGG.................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................ccACTATCCACCAGCGC................................................... | 17 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................TCCAGGGTAGCTGAGGG..................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |