| (1) B-CELL | (1) BRAIN | (1) CELL-LINE | (1) FIBROBLAST | (1) HEART | (9) LIVER | (3) OTHER | (1) PLACENTA | (2) SKIN |
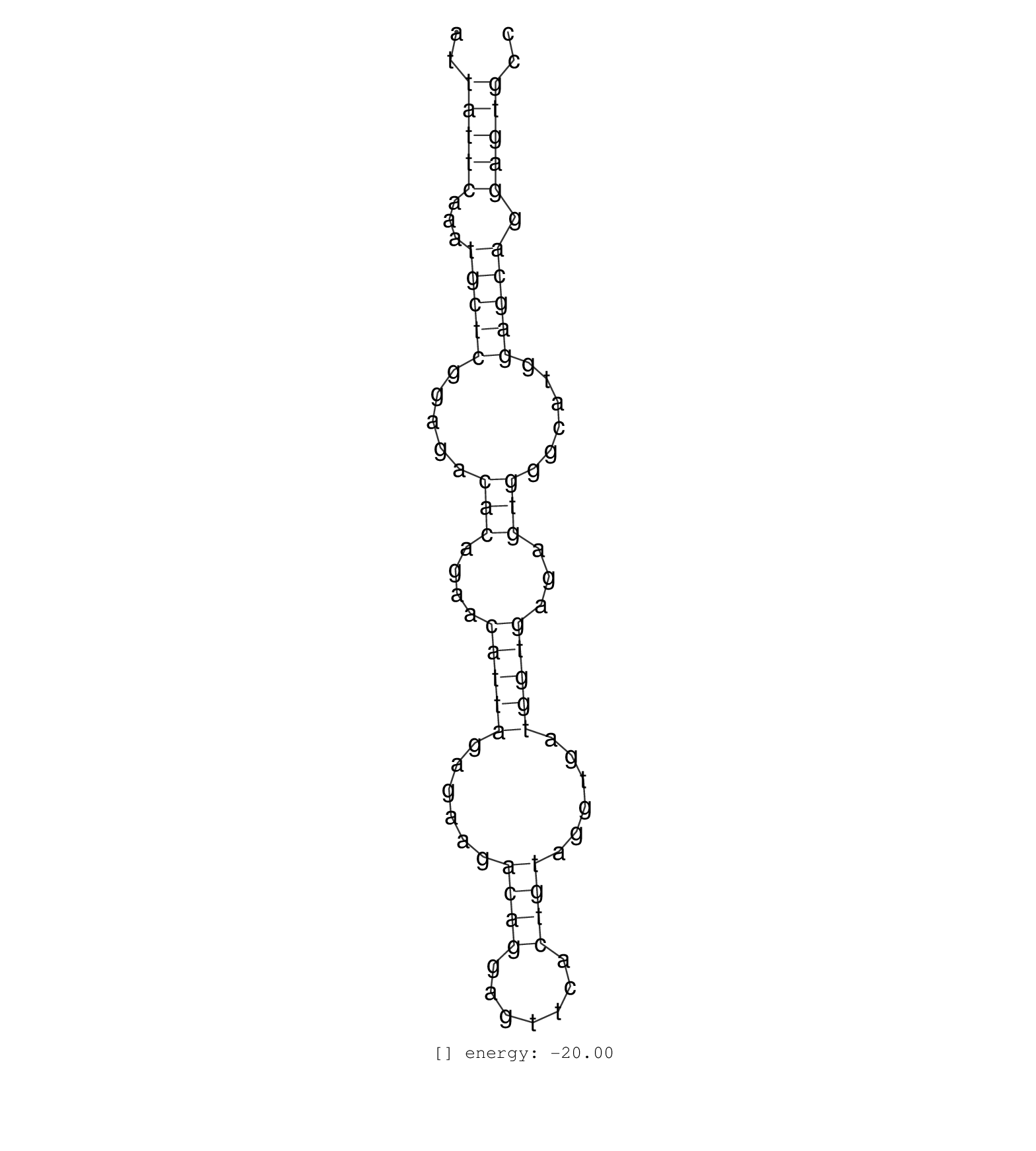
| CCAAGGGAGATGGAGGTTTGGGGTGGGAGAATGTGTTGGATTCCAGCTATATTATTCAAATGCTCGGAGACACAGAACATTAGAGAAGACAGGAGTTCACTGTAGGTGATGGTGAGAGTGGGCATGGAGCAGGAGTGCCAAGAGATGAAGTTATAAAACTAAGCAGGAGTTAGATCATGAAGAACTTTA ....................................................(((((...(((((.....(((....(((((......((((.......))))......)))))...)))......))))).))))).................................................... ..................................................51......................................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189784 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR343335 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................TGGGGTGGGAGAATGgagt........................................................................................................................................................ | 19 | GAGT | 45.00 | 0.00 | 14.00 | 12.00 | 9.00 | 3.00 | 1.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgaga........................................................................................................................................................ | 19 | GAGA | 19.00 | 0.00 | 6.00 | 5.00 | 2.00 | 2.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgggt........................................................................................................................................................ | 19 | GGGT | 13.00 | 0.00 | 6.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................TGGGGTGGGAGAATGgata........................................................................................................................................................ | 19 | GATA | 5.00 | 0.00 | 2.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgag......................................................................................................................................................... | 18 | GAG | 3.00 | 0.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgtga........................................................................................................................................................ | 19 | GTGA | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgtgg........................................................................................................................................................ | 19 | GTGG | 2.00 | 0.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgaaa........................................................................................................................................................ | 19 | GAAA | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTtgtt....................................................................................................................................................... | 20 | TGTT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGaaaa........................................................................................................................................................ | 19 | AAAA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTGTTtt...................................................................................................................................................... | 21 | TT | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGggtt........................................................................................................................................................ | 19 | GGTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTGgttt...................................................................................................................................................... | 21 | GTTT | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTagt........................................................................................................................................................ | 19 | AGT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTGTTtga..................................................................................................................................................... | 22 | TGA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTGgggt...................................................................................................................................................... | 21 | GGGT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GGGGTGGGAGAATGTat......................................................................................................................................................... | 17 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................AGAGAAGACAGGAGTcagg......................................................................................... | 19 | CAGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................GGGTGGGAGAATGTGTattt..................................................................................................................................................... | 20 | ATTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................TGGGAGAATGTGTTGaata................................................................................................................................................... | 19 | AATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................TGGAGCAGGAGTGCCAtcg.............................................. | 19 | TCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgaat........................................................................................................................................................ | 19 | GAAT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTaga........................................................................................................................................................ | 19 | AGA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATCATGAAGAACTTTA | 18 | 2 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTatag....................................................................................................................................................... | 20 | ATAG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGgatg........................................................................................................................................................ | 19 | GATG | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGGGTGGGAGAATGTat......................................................................................................................................................... | 18 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................TCGGAGACACAGAAC............................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| CCAAGGGAGATGGAGGTTTGGGGTGGGAGAATGTGTTGGATTCCAGCTATATTATTCAAATGCTCGGAGACACAGAACATTAGAGAAGACAGGAGTTCACTGTAGGTGATGGTGAGAGTGGGCATGGAGCAGGAGTGCCAAGAGATGAAGTTATAAAACTAAGCAGGAGTTAGATCATGAAGAACTTTA ....................................................(((((...(((((.....(((....(((((......((((.......))))......)))))...)))......))))).))))).................................................... ..................................................51......................................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189784 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR037937(GSM510475) 293cand2. (cell line) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR343335 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................atTGATGGTGAGAGTGGGCA................................................................. | 20 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................agCTGTAGGTGATGGTGA.......................................................................... | 18 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| gtAAGGGAGATGGAGGTT........................................................................................................................................................................... | 18 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................cctAGTGCCAAGAGATGA......................................... | 18 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |