| (2) AGO2.ip | (4) B-CELL | (1) BRAIN | (9) BREAST | (16) CELL-LINE | (1) CERVIX | (4) HEART | (2) HELA | (3) LIVER | (1) OTHER | (7) SKIN | (1) UTERUS |
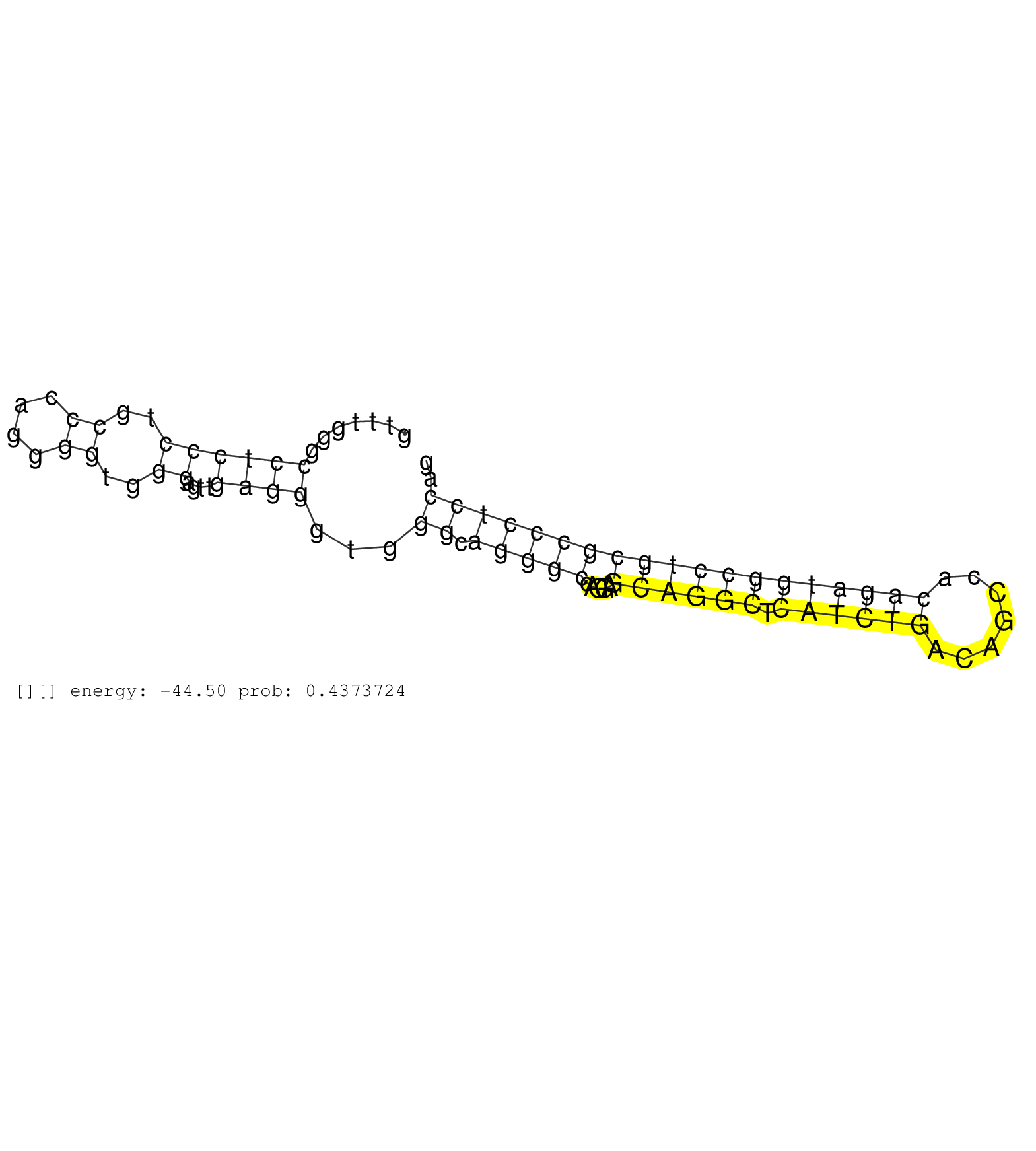
| GGCGTCACCATGCCGAATGCCTGCGTCCACGCTGCCAAGCCTGTGACGAGGTTTGGGCCTCCCTGCCCAGGGGTGGGAGTTGAGGGTGGGCAGGGCCAGGAGCAGGCTCATCTGACAGCCACAGATGGCCTGCGCCCTCCAGATCATCTTCTCCCCTGAGTGCACGGAGGCTGAGGGCCGCCACTGGCACAT .............................................................((..((....))..)).((((.((((((((...((.....))..))))))))..))))......................................................................... ...........................................................60.............................................................123................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR038863(GSM458546) MM603. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR038857(GSM458540) D20. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR191544(GSM715654) 126genomic small RNA (size selected RNA from . (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR038855(GSM458538) D10. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR040024(GSM532909) G613N. (cervix) | TAX577453(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR553574(SRX182780) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................AGGAGCAGGCTCATCTGACAGC......................................................................... | 22 | 1 | 10.00 | 10.00 | 2.00 | - | 1.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACAGCC........................................................................ | 24 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAGCC........................................................................ | 23 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAGCCA....................................................................... | 24 | 1 | 6.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................CCAGGAGCAGGCTCATCTGACAGCC........................................................................ | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACAGC......................................................................... | 23 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAGCA........................................................................ | 23 | 1 | 2.00 | 10.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAGA......................................................................... | 22 | 1 | 2.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACAG.......................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCAGGAGCAGGCTCATCTGACA........................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACAA.......................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................AGGAGCAGGCTCATCTGACAGCCACAGA................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACATC......................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............GAATGCCTGCGTCCACGCT............................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAGCCACAG.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGGAGTTGAGGGTGATTT.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................CAGGAGCAGGCTCATCTGACAGA......................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCAGGAGCAGGCTCATCTG.............................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................CAAGCCTGTGACGAGA............................................................................................................................................. | 16 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................CCAGGAGCAGGCTCATCTGA............................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGACAG.......................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................TGCCAAGCCTGTGACGAG.............................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................CAGGAGCAGGCTCATCTGACAGCCAT...................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................AGGCTGAGGGCCGCCACTGGCACACGG | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................CAGGAGCAGGCTCATCTGCCAG.......................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................AGGAGCAGGCTCATCTGACA........................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGGAGCAGGCTCATCTGCG............................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ................................................................GCCCAGGGGTGGGAGTGGCA............................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................CCAGGAGCAGGCTCATCTGACAGC......................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CGGAGGCTGAGGGCCG............ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGCACGGAGGCTGAGAGC.............. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ............................................................................................GGGCCAGGAGCAGGCTCATCTGACA........................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGGAGCAGGCTCATCTGA............................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .................................................................................................AGGAGCAGGCTCATCTGATT........................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGCGTCACCATGCCGAATGCCTGCGTCCACGCTGCCAAGCCTGTGACGAGGTTTGGGCCTCCCTGCCCAGGGGTGGGAGTTGAGGGTGGGCAGGGCCAGGAGCAGGCTCATCTGACAGCCACAGATGGCCTGCGCCCTCCAGATCATCTTCTCCCCTGAGTGCACGGAGGCTGAGGGCCGCCACTGGCACAT .............................................................((..((....))..)).((((.((((((((...((.....))..))))))))..))))......................................................................... ...........................................................60.............................................................123................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR038863(GSM458546) MM603. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR038857(GSM458540) D20. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR191544(GSM715654) 126genomic small RNA (size selected RNA from . (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR038855(GSM458538) D10. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR040024(GSM532909) G613N. (cervix) | TAX577453(Rovira) total RNA. (breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR553574(SRX182780) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................CCTCCAGATCATCTTCTAT...................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................TGGGCCTCCCTGCCCAGGG........................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....TCACCATGCCGAATGCCTGCGTCCACGCTGA............................................................................................................................................................. | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |