| (1) BREAST | (20) CELL-LINE | (2) FIBROBLAST | (1) HELA | (1) OTHER | (15) SKIN | (1) TESTES | (1) XRN.ip |
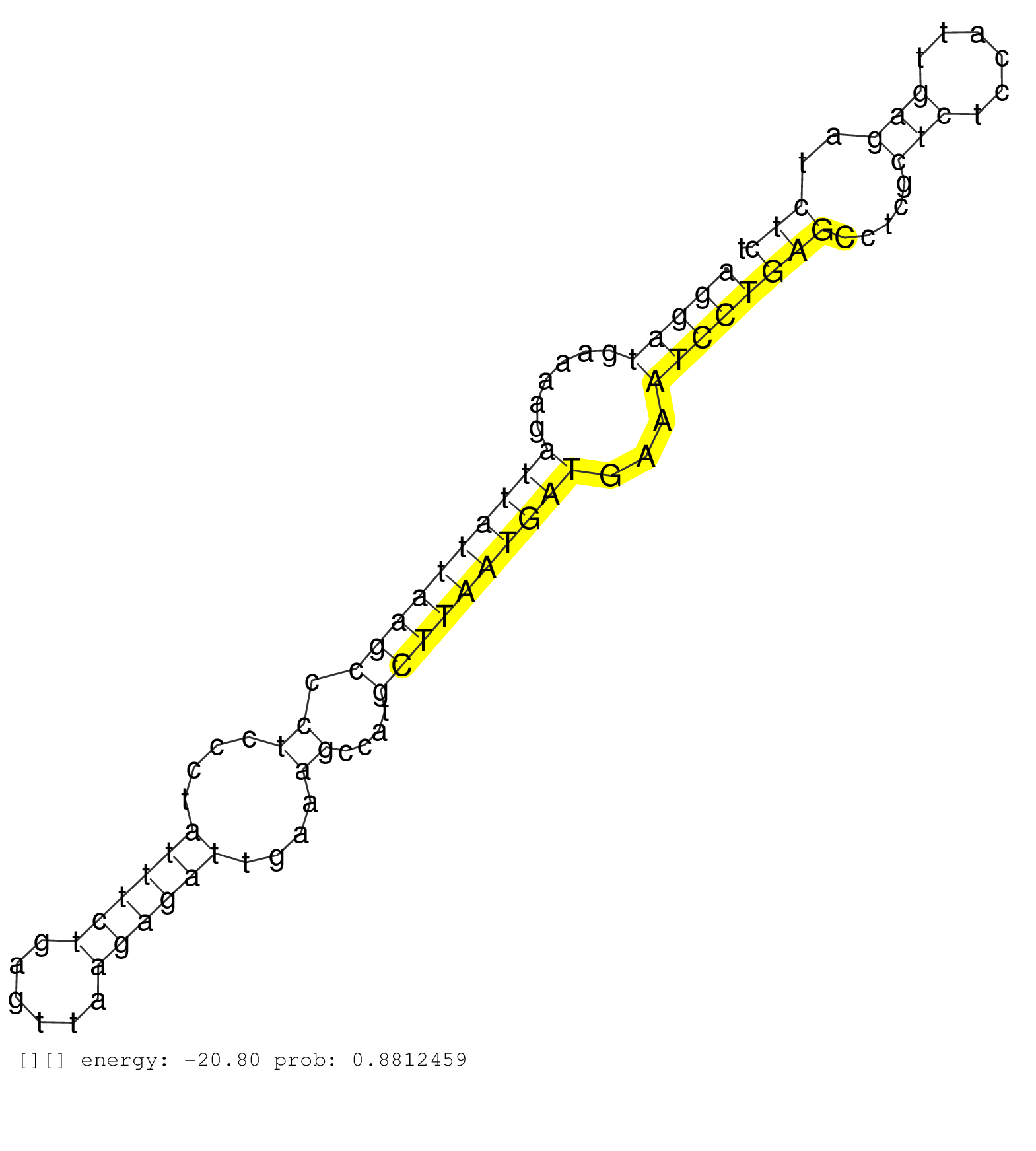
| CTGTGACATTACCCAAGTAGCCTCAAATCCTTAGACCTACAGTGAAGCAGGTAAATTCCCAGGCATGCTAAACCCTTAATAGGATTAGCTCCCTTAAGTCAGGATGAAAAGATTATTAAGCCCTCCCTATTTCTGAGTTAAGAGATTGAAAGCCATGCTTAATGATGAAATCCTGAGCCTCGCTCTCCATTGAGATCTCTGCTGGTGACTTGGAGTGCTGTAGATGGGAGTGATACGTCTCCGAGTGAAT ..................................................................................................(((((((......((((((((((.((....((((((......))))))....))....))))))))))...))))))).......................................................................... .........................................................................................90..........................................................................................182.................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191556(GSM715666) 45genomic small RNA (size selected RNA from t. (breast) | GSM416733(GSM416733) HEK293. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................CTTAATGATGAAATCCTGAGC........................................................................ | 21 | 1 | 145.00 | 145.00 | 40.00 | 32.00 | 20.00 | 11.00 | 10.00 | 4.00 | 3.00 | - | 3.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | 2.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................................ACTTGGAGTGCTGTAGGCTT....................... | 20 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................GCTTAATGATGAAATCCTGAGC........................................................................ | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................CTTGGAGTGCTGTAGGCTT....................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................CTTAATGATGAAATCCTG........................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TTAATGATGAAATCCTGAGC........................................................................ | 20 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CTTGGAGTGCTGTAGGCT........................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ...............................................................................................................................................................................................................ACTTGGAGTGCTGTAGGCT........................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TGAGTTAAGAGATTGAATAG................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................GTGAAGCAGGTAAATACGT.............................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................AGTGCTGTAGATGGGAGTGGGGC.............. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................GAGTGCTGTAGATGGGAGTGATAC.............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................CTTAATGATGAAATCCTGAG......................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................CTTAATGATGAAATCCTTATC........................................................................ | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................TAAGTCAGGATGAAAAAAC......................................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................ACTTGGAGTGCTGTAGGCA........................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................GGAGTGCTGTAGATGTCTG.................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................CTTAATGATGAAATCCTGCGC........................................................................ | 21 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................CTTAATGATGAAATCCTTAGC........................................................................ | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TGAGATCTCTGCTGGTGACTTGGAGTGC................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GTGCTGTAGATGGGAGACC................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TGAGTTAAGAGATTGAAA................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CCCAAGTAGCCTCAA................................................................................................................................................................................................................................ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| CTGTGACATTACCCAAGTAGCCTCAAATCCTTAGACCTACAGTGAAGCAGGTAAATTCCCAGGCATGCTAAACCCTTAATAGGATTAGCTCCCTTAAGTCAGGATGAAAAGATTATTAAGCCCTCCCTATTTCTGAGTTAAGAGATTGAAAGCCATGCTTAATGATGAAATCCTGAGCCTCGCTCTCCATTGAGATCTCTGCTGGTGACTTGGAGTGCTGTAGATGGGAGTGATACGTCTCCGAGTGAAT ..................................................................................................(((((((......((((((((((.((....((((((......))))))....))....))))))))))...))))))).......................................................................... .........................................................................................90..........................................................................................182.................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR207116(GSM721078) Nuclear RNA. (cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | SRR029129(GSM416758) SW480. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR191556(GSM715666) 45genomic small RNA (size selected RNA from t. (breast) | GSM416733(GSM416733) HEK293. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................ATGAAATCCTGAGCCTCC.................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |