| (2) BRAIN | (15) BREAST | (4) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (1) HEART | (4) LIVER | (3) OTHER | (1) RRP40.ip |
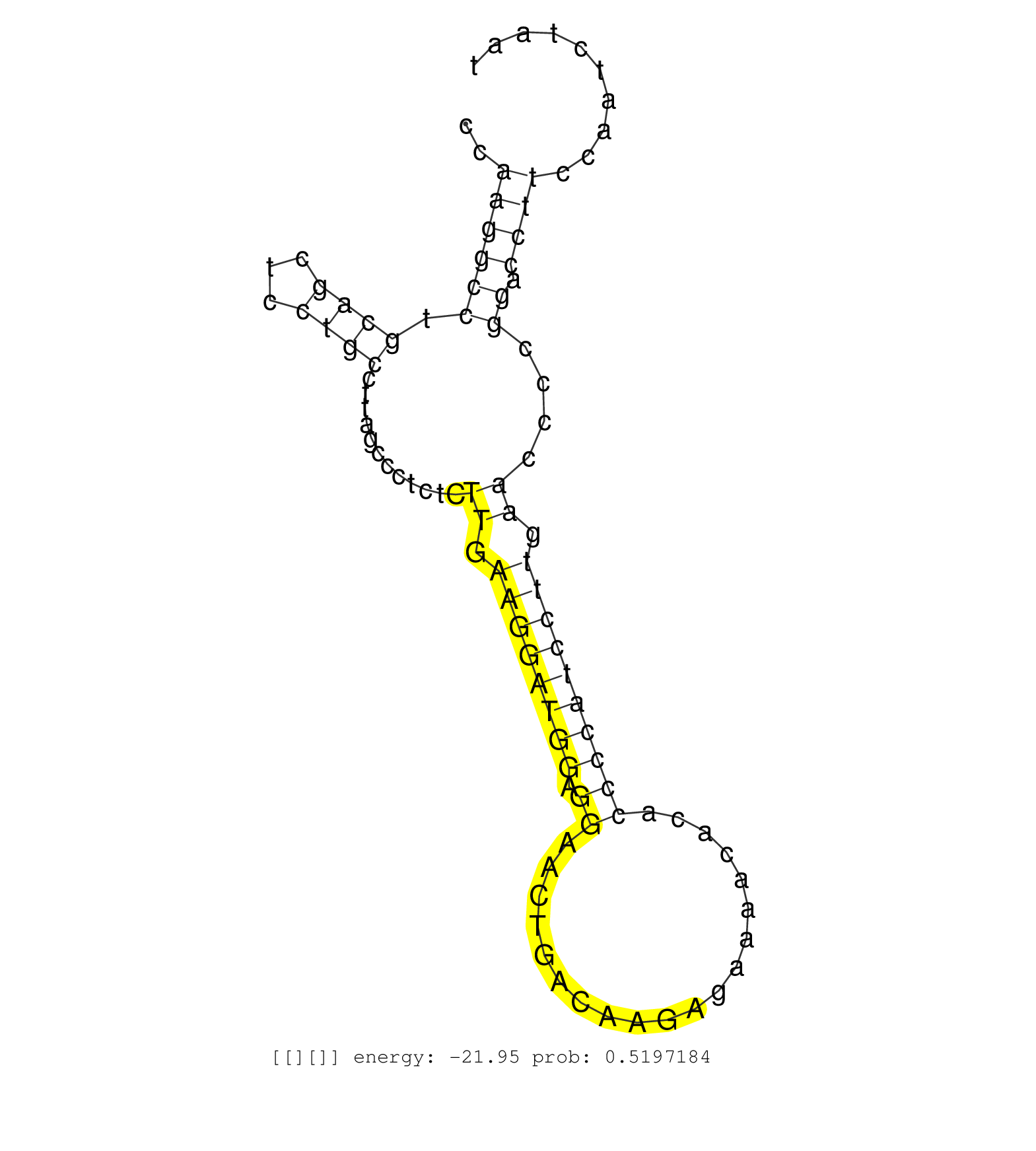
| CACACCACATCCACAGACCTCAAGTCAGGGCCCCTGAACTGTTTAAAGTTGGCTCTCTGAGCACATTGACAGGGCCCACAGCAGAGGACGCTGCTGCCCAGGCCCTGGAGTCTATCCCACTAGGATTGGCAGGAGGCAGGAGGACACAGCCAGAAGGTGAGCAGATTTGTATGACCAAGTTACCCCATTGCCATCCACAGGGAAATGTTCCACTGCAGCCGAGTGCCCGTGTGTCTGTGTCTGAGAAGAA .....................................................(((((..((.......(((((((..((((((......))))))....)))))))..(((.((((.....)))).))).....)).)))))........................................................................................................... ..................................................51................................................................................................149................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | TAX577453(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189787 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | TAX577739(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189785 | TAX577590(Rovira) total RNA. (breast) | TAX577742(Rovira) total RNA. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | GSM532887(GSM532887) G761N. (cervix) | TAX577738(Rovira) total RNA. (breast) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................AGCACATTGACAGGGATTT............................................................................................................................................................................ | 19 | 30.00 | 0.00 | 2.00 | 4.00 | 6.00 | 2.00 | 5.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATTA............................................................................................................................................................................ | 19 | 8.00 | 0.00 | 1.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATT............................................................................................................................................................................. | 18 | 6.00 | 0.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGTTTT............................................................................................................................................................................ | 19 | 4.00 | 0.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGAATT............................................................................................................................................................................ | 19 | 3.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................GTGTGTCTGTGTCTGAGAAGAA | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................AGCACATTGACAGGGAATA............................................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...........................................................AGCACATTGACAGGGCTTT............................................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATGT............................................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGAT.............................................................................................................................................................................. | 17 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGAGCT............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGCTTTC........................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................GGCAGGAGGACACAGCGGAC................................................................................................ | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGACT............................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGTA.............................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGTTTC............................................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATTG............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGTTTA............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATAA............................................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATGG............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGTCTA............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................GCAGGAGGCAGGAGGATCGG...................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGCTGT............................................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGAGTT............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGCACATTGACAGGGATAG............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................CAGGAGGCAGGAGGACACTC..................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ........................TCAGGGCCCCTGAACTGTTT.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CAGGAGGCAGGAGGACA........................................................................................................ | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| CACACCACATCCACAGACCTCAAGTCAGGGCCCCTGAACTGTTTAAAGTTGGCTCTCTGAGCACATTGACAGGGCCCACAGCAGAGGACGCTGCTGCCCAGGCCCTGGAGTCTATCCCACTAGGATTGGCAGGAGGCAGGAGGACACAGCCAGAAGGTGAGCAGATTTGTATGACCAAGTTACCCCATTGCCATCCACAGGGAAATGTTCCACTGCAGCCGAGTGCCCGTGTGTCTGTGTCTGAGAAGAA .....................................................(((((..((.......(((((((..((((((......))))))....)))))))..(((.((((.....)))).))).....)).)))))........................................................................................................... ..................................................51................................................................................................149................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | TAX577453(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577740(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | TAX577580(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR189787 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | TAX577739(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577588(Rovira) total RNA. (breast) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR191566(GSM715676) 94genomic small RNA (size selected RNA from t. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189785 | TAX577590(Rovira) total RNA. (breast) | TAX577742(Rovira) total RNA. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | GSM532887(GSM532887) G761N. (cervix) | TAX577738(Rovira) total RNA. (breast) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................TGCCATCCACAGGGAGAC............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................TGTATGACCAAGTTATG.................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GGCTCTCTGAGCACATTGGAA................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ......................................................................................GACGCTGCTGCCCAGGCA.................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................GCTGCCCAGGCCCTGTCAG........................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |