| (1) B-CELL | (2) CELL-LINE | (2) HEART | (14) LIVER | (4) OTHER | (6) SKIN | (1) TESTES |
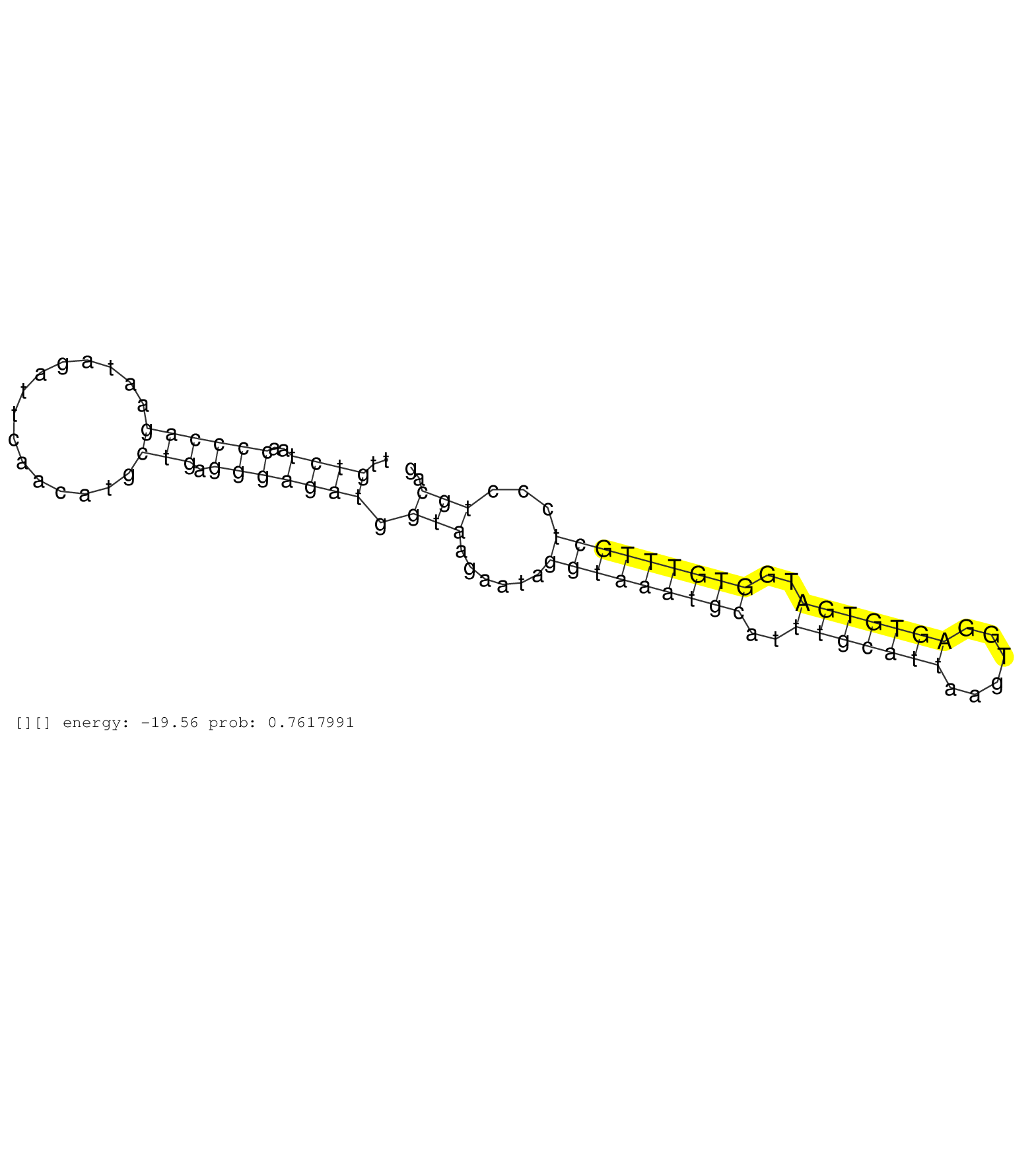
| TCAACTTTCTCACTCTTAAGTATTTTAATAGCCCCGATTCTTCCTTTACCATTAAAAGCCAAAATTGTTTTTATTAAACATAGCAGATAAGTCTGGGAGCTTGTCTAACCCCAGAATAGATTCAACATGCTGAGGGAGATGGTAAGAATAGGTAAATGCATTTGCATTAAGTGGAGTGTGATGGTGTTTGCTCCCTGCAGGTATGGTTTCTGTGGATTCTCTGCTCTTTGTGCTCTCCTCCCCACATTGG ......................................................................................................................................................(((((((((..(((((((......)))))))..))))))))).......................................................... ................................................................................................................................129..............................................................194...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553576(SRX182782) source: Testis. (testes) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343334 | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR343335 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTG............................................................ | 19 | 1 | 54.00 | 54.00 | 9.00 | 8.00 | 12.00 | 5.00 | 4.00 | 2.00 | 4.00 | 2.00 | 1.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTGA........................................................... | 20 | 1 | 25.00 | 54.00 | 5.00 | 3.00 | 1.00 | 2.00 | 1.00 | 1.00 | 1.00 | 4.00 | 2.00 | - | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTGT........................................................... | 20 | 1 | 17.00 | 54.00 | 2.00 | 6.00 | 1.00 | 4.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTT............................................................. | 18 | 1 | 15.00 | 15.00 | 1.00 | - | 1.00 | 1.00 | 2.00 | 3.00 | - | 1.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGGTGG............................................................ | 19 | 3.00 | 0.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGGTGT............................................................ | 19 | 2.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................GGGAGATGGTAAGAATTTTT................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................ATTTGCATTAAGTGGAACT........................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGCTTG............................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................CAGAATAGATTCAACTGT......................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTAAT.......................................................... | 21 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTGTT.......................................................... | 21 | 1 | 1.00 | 54.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GGAGTGTGATGGTGTTTGT........................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTAA........................................................... | 20 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTT.............................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTGTT........................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TAAGTGGAGTGTGATGGTG................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGGAGTGTGATGGTGTTTA............................................................ | 19 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGGTATGGTTTCTGTGGATTCTA............................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................TGCAGGTATGGTTTCTGTGGATTCTCTG........................... | 28 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| .............................................................................................................................................................................................................GTTTCTGTGGATTCTCTGCTCTTTGT................... | 26 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CTGTGGATTCTCTGCTC........................ | 17 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| .........................................................................................................................................................................................................TATGGTTTCTGTGGATTCTCTG........................... | 22 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| .....................................................................................................................................................................................................CAGGTATGGTTTCTGTGGATTCTCTGC.......................... | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TCTCCTCCCCACATTG. | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| TCAACTTTCTCACTCTTAAGTATTTTAATAGCCCCGATTCTTCCTTTACCATTAAAAGCCAAAATTGTTTTTATTAAACATAGCAGATAAGTCTGGGAGCTTGTCTAACCCCAGAATAGATTCAACATGCTGAGGGAGATGGTAAGAATAGGTAAATGCATTTGCATTAAGTGGAGTGTGATGGTGTTTGCTCCCTGCAGGTATGGTTTCTGTGGATTCTCTGCTCTTTGTGCTCTCCTCCCCACATTGG ......................................................................................................................................................(((((((((..(((((((......)))))))..))))))))).......................................................... ................................................................................................................................129..............................................................194...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553576(SRX182782) source: Testis. (testes) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343334 | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR343335 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TTGTTTTTATTAAACAAGC....................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................AGTGGAGTGTGATGGCTGA.............................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .............................AGCCCCGATTCTTCCGAGA.......................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................................................AGTGGAGTGTGATGGCTG............................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................TGTCTAACCCCAGAAGTAG.................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................GTGCTCTCCTCCCCACCAGG. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |