| (8) B-CELL | (13) BREAST | (10) CELL-LINE | (2) CERVIX | (3) HEART | (6) LIVER | (1) OTHER | (27) SKIN | (2) UTERUS |
| CACAGGGCAGTGATTTAGGGAGACACACTAGCTCTAGAATCAGACACATGTGGATTCATGTGCCCACTCTGCCCCTTACGGGCTCCGCTGGCCTCGGGCGAGTCATAGCCTCTCTGAACCTCCCTTTCTTCTTTGGTAAAATTGGGGTCATCAATGCCTGCCCCCCCTCCCAGCTGTGGTACTGACACTCTTCTCCCCAGGGACAGAGCTGCCCAGCCCTCCGTCTGTGTGGTTTGAAGCAGAATTTTTC ...............................................................................(((....(((((....((((.((.(((.(((((.....(((..............)))......))))).....))).))))))......))))).(((.......))).......))).................................................... ...........................................................................76..........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | SRR191569(GSM715679) 54genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR191442(GSM715552) 107genomic small RNA (size selected RNA from . (breast) | SRR029128(GSM416757) H520. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191559(GSM715669) 65genomic small RNA (size selected RNA from t. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR191455(GSM715565) 181genomic small RNA (size selected RNA from . (breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR040015(GSM532900) G623T. (cervix) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................GGTACTGACACTCTTCTCCCCAG.................................................. | 23 | 1 | 57.00 | 57.00 | 10.00 | 4.00 | 4.00 | 5.00 | 1.00 | 3.00 | 2.00 | 2.00 | 1.00 | - | 1.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - |
| ................................................................................................................................................................................TGGTACTGACACTCTTCTCCCCAG.................................................. | 24 | 1 | 52.00 | 52.00 | 2.00 | 6.00 | 6.00 | - | 5.00 | 2.00 | 3.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................GTGGTACTGACACTCTTCTCCCCAG.................................................. | 25 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGTACTGACACTCTTCTCCCCAGA................................................. | 25 | 1 | 4.00 | 52.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGTACTGACACTCTTCTCCCCAGAA................................................ | 26 | 1 | 3.00 | 52.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CTGTGGTACTGACACTCTTCTCCCCAG.................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CAGCTGTGGTACTGACACTCTTCTCCCCAG.................................................. | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GGTACTGACACTCTTCTCCCCAT.................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGTGGTACTGACACTCTTCTCCCCAG.................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGTGGTACTGACACTCTTCTCCCCCG.................................................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................CCCAGGGACAGAGCTGAA..................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................CTGTGGTACTGACACTCTTCTCCCCAGA................................................. | 28 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......GCAGTGATTTAGGGAGA................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTGGTACTGACACTCTTCTCCCCCGA................................................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................CCAGCTGTGGTACTGACACTCTTCTCCCCAG.................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CAGGGCAGTGATTTAGGGAGACA................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GGCTCCGCTGGCCTCGGGT....................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CCCCCCCTCCCAGCTGTGGTACTGACACTC............................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGTACTGACACTCTTCTCCCCAGGGAC.............................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CTAGCTCTAGAATCAGA.............................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CTCCGTCTGTGTGGTTTGAAGCAG........ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GCCTCGGGCGAGTCATAGCCT........................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCATAGCCTCTCTGAACCTTGTT............................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................TCTGTGTGGTTTGAAGCAGAA...... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GGTACTGACACTCTTCTCCCCAGAA................................................ | 25 | 1 | 1.00 | 57.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GGTACTGACACTCTTTTCC...................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GGTACTGACACTCTTCTCCCCGG.................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
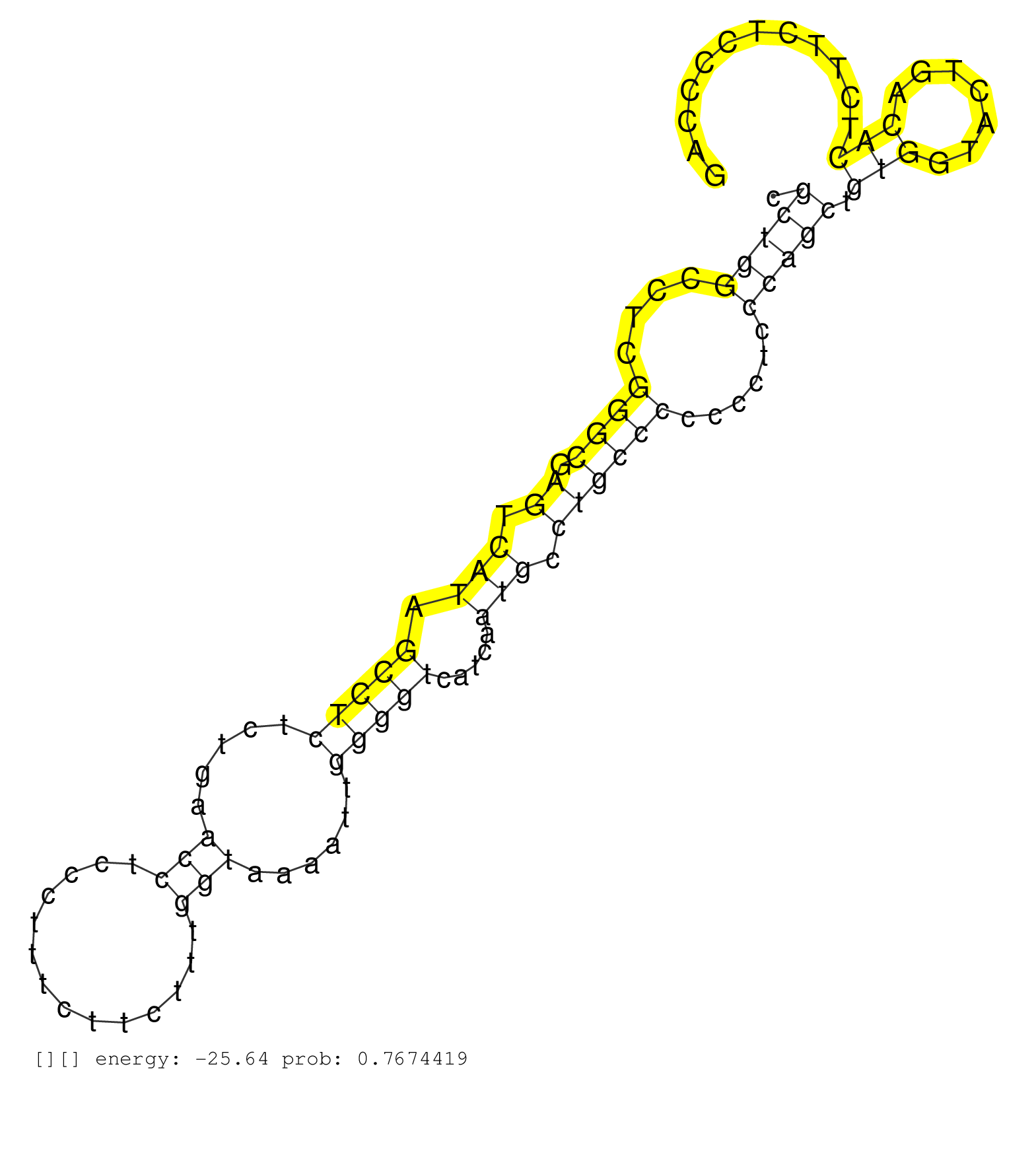
| CACAGGGCAGTGATTTAGGGAGACACACTAGCTCTAGAATCAGACACATGTGGATTCATGTGCCCACTCTGCCCCTTACGGGCTCCGCTGGCCTCGGGCGAGTCATAGCCTCTCTGAACCTCCCTTTCTTCTTTGGTAAAATTGGGGTCATCAATGCCTGCCCCCCCTCCCAGCTGTGGTACTGACACTCTTCTCCCCAGGGACAGAGCTGCCCAGCCCTCCGTCTGTGTGGTTTGAAGCAGAATTTTTC ...............................................................................(((....(((((....((((.((.(((.(((((.....(((..............)))......))))).....))).))))))......))))).(((.......))).......))).................................................... ...........................................................................76..........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | TAX577580(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | SRR191569(GSM715679) 54genomic small RNA (size selected RNA from t. (breast) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR191442(GSM715552) 107genomic small RNA (size selected RNA from . (breast) | SRR029128(GSM416757) H520. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191559(GSM715669) 65genomic small RNA (size selected RNA from t. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR191455(GSM715565) 181genomic small RNA (size selected RNA from . (breast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR040015(GSM532900) G623T. (cervix) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................CAGCCCTCCGTCTGTGT.................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................AGAGTGGGCACATGAATC.................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CATAGCCTCTCTGAAGTAC................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................GGAGACACACTAGCTATGA..................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CAGAGCTGCCCAGCCCTGA............................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ................................................................................................................TCTGAACCTCCCTTTCTCC....................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................ACACTCTTCTCCCCAGGGGGC............................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................GCAGCTCTGTCCCTGGG...................................... | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GGAGGGGGGGCAGGC................................................................................ | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |