| (1) AGO2.ip | (2) BRAIN | (1) BREAST | (12) CELL-LINE | (2) HEART | (1) HELA | (4) LIVER | (2) OTHER | (9) SKIN | (1) XRN.ip |
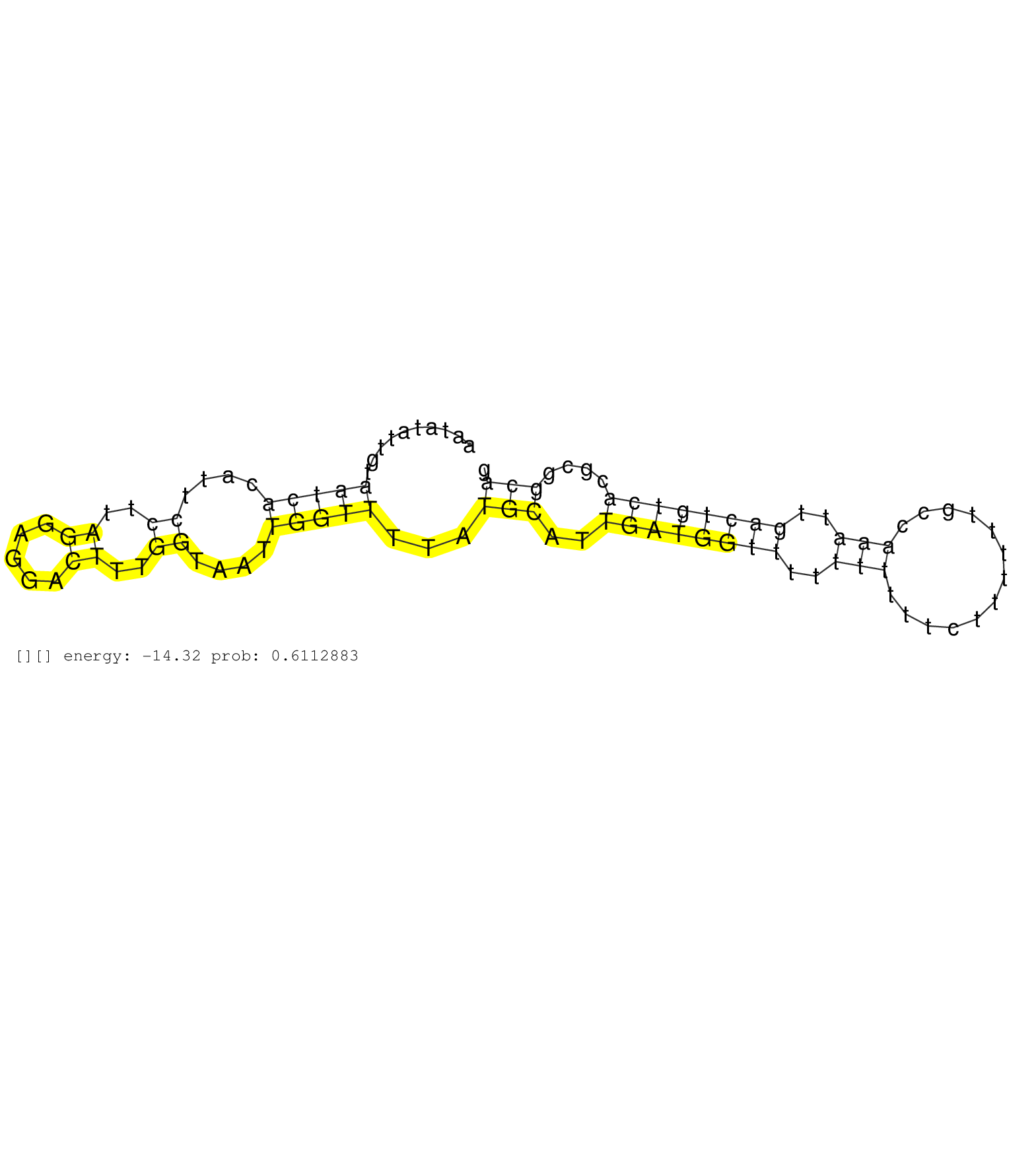
| CGTTAGAAACACACAGCAGTGTTCAGGTGTGGGCAGGTAGACACACCTGGGGTGGTCCCGCAGACTGCATGGAGATGGCAGTGGCTCTCCGTGGCATCACTGCTGCATGACAGTGTGACTTTCATGTGGATCTGTGTTGCTGCAACGGAAGTCTTTGGCTGGTAGAATTTCCACATGGATTTTTTTTTTCTCTTAAAAAGCAAGAGAACAAACTGCGATTGAGCCTCTTTTTAGCGCTCAGTAGAGCAGG .........................................................................................................................................................(((((..((.((((..(((....)))..))))...))..)))))..................................................... ......................................................................................................................................................151..............................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR207110(GSM721072) Nuclear RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TTTTTTTTTCTCTTAACG.................................................... | 18 | 8.00 | 0.00 | - | - | - | 2.00 | 1.00 | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTTTTTTTTTCTCTTAACG.................................................... | 19 | 5.00 | 0.00 | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGCAACGGAAGTCTTTGGCTGGTAGAATTCTG.............................................................................. | 32 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GATTTTTTTTTTCTCTTAAAGAG.................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................TGGCTGGTAGAATTTATG............................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................CTGGTAGAATTTCCACATGG........................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GTGACTTTCATGTGGATCTGTGTTGCTG............................................................................................................ | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GAGATGGCAGTGGCTCTCCGTGG............................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................................ATGGATTTTTTTTTTCTCCC........................................................ | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................AAAGCAAGAGAACAAACTGCGATTG............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................ACTTTCATGTGGATCTGTGTTGCTGCAACG....................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GAGATGGCAGTGGCTCTCCGTGGCAT......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TTGCTGCAACGGAAGTCTTTGGCT.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AACGGAAGTCTTTGGCTGGTA...................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGGTAGAATTTCCACATGGAT...................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGATGGCAGTGGCTCTCCGTGGGA.......................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................TTCATGTGGATCTGTGTTGCTGCAACG....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GCAACGGAAGTCTTTGGCTGGTAGAA................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CAGACTGCATGGAGATG............................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................AACGGAAGTCTTTGGCTGGTAGAA................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................ACTTTCATGTGGATCTGTG.................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................AAGTCTTTGGCTGGTAGAATTTCCACATGGA....................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............GCAGTGTTCAGGTGTGGGCAGGTAGACACAC............................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTTTTTTTTCTCTTAAAAAATG................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................CTGCAACGGAAGTCTTTGGCTGGTAGAATTTC............................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GGTAGAATTTCCACATGGAT...................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTTTTTTTTCTCTTAACAGG.................................................. | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................AACGGAAGTCTTTGGCTG......................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AGAGAACAAACTGCGATTG............................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGACTTTCATGTGGATCTGTGTTGC.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................TGGGCAGGTAGACACTTTG.......................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................CAGTGTGACTTTCATGTGGATCTGTGTT................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GTGGATCTGTGTTGCTGCAACG....................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGCAACGGAAGTCTTTGGCTGG........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGGGGTGGTCCCGCAGACTGTATG................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................TGGATCTGTGTTGCTGCAACGGAAGGC................................................................................................. | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................GCTGCAACGGAAGTCTT............................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGATCTGTGTTGCTGCAACGGAAGTCTTTGGC........................................................................................... | 33 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................GAGAACAAACTGCGATTGA............................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................ACACCTGGGGTGGTCCCGCAGACTGCACG................................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................TTCATGTGGATCTGTGTTGC.............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTTTTTTTTCTCTTAAAAAATG................................................ | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................ACGGAAGTCTTTGGCTGGTAGAATCTCC.............................................................................. | 28 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................TTGGCTGGTAGAATTTCCACATGGATTT.................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TGTTCAGGTGTGGGCAGGTAGACA............................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGATGGCAGTGGCTCTCCGTGGCA.......................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CTGCAACGGAAGTCTTTGGCTGGTAGA.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TTCATGTGGATCTGTGTTGCTGC........................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................ACATGGATTTTTTTTGC............................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................................................................TTTTCTCTTAAAAAGCAA............................................... | 18 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CTGCATGGAGATGGC........................................................................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| CGTTAGAAACACACAGCAGTGTTCAGGTGTGGGCAGGTAGACACACCTGGGGTGGTCCCGCAGACTGCATGGAGATGGCAGTGGCTCTCCGTGGCATCACTGCTGCATGACAGTGTGACTTTCATGTGGATCTGTGTTGCTGCAACGGAAGTCTTTGGCTGGTAGAATTTCCACATGGATTTTTTTTTTCTCTTAAAAAGCAAGAGAACAAACTGCGATTGAGCCTCTTTTTAGCGCTCAGTAGAGCAGG .........................................................................................................................................................(((((..((.((((..(((....)))..))))...))..)))))..................................................... ......................................................................................................................................................151..............................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR207110(GSM721072) Nuclear RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|