| (1) AGO2.ip | (7) B-CELL | (1) BRAIN | (2) BREAST | (14) CELL-LINE | (2) CERVIX | (1) HELA | (2) LIVER | (1) OTHER | (3) SKIN |
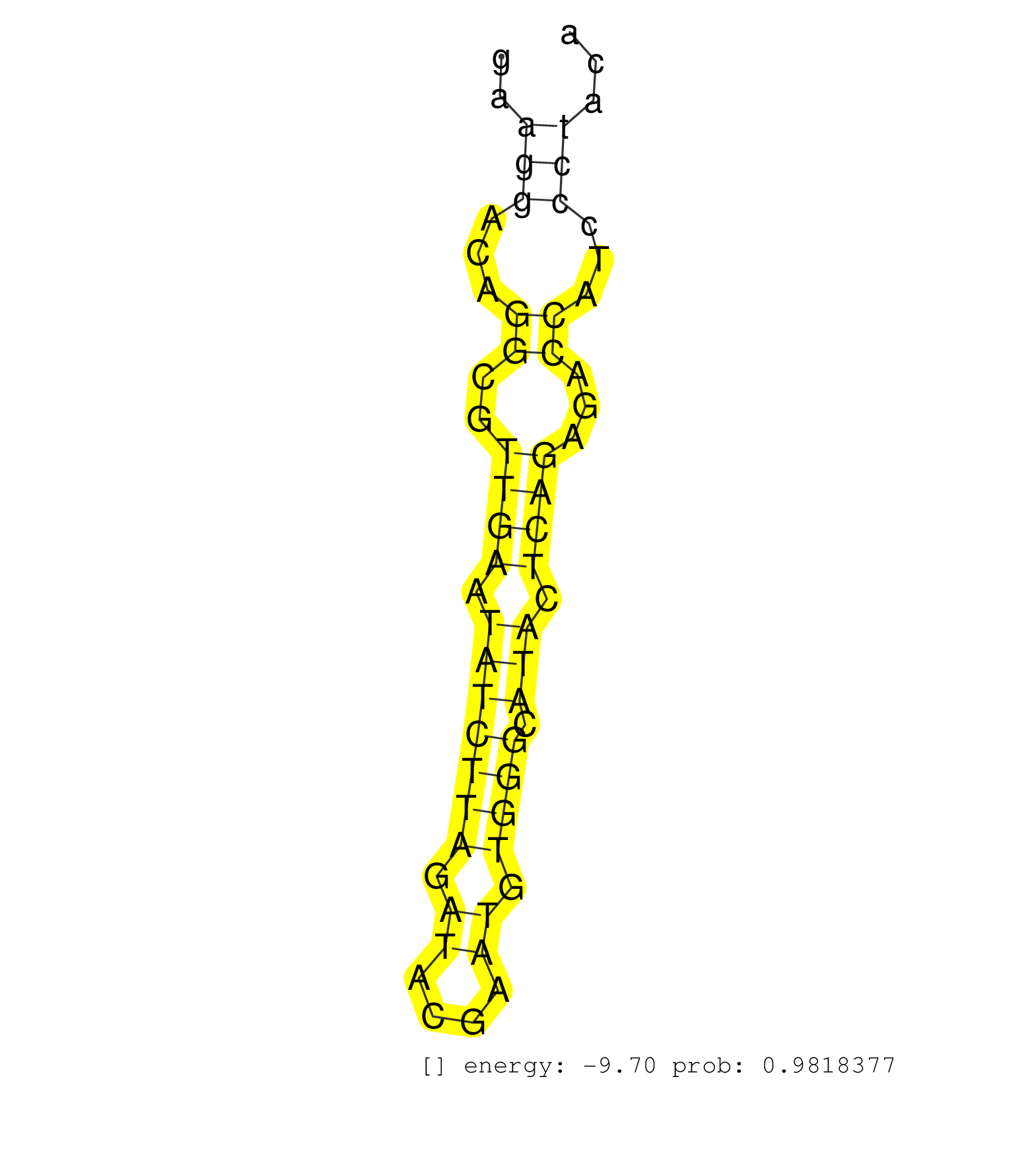
| TTTGCCTTTGTGTTTCCAACAGGTCAGCGGACAGATTGCAGTCTAGCCAGGTGAGTGTGGCCTGGGACAGGCCTGGGAAGTGATAGAGGCTTGGTGGGTAGAGCAATCTTACTCTGGGAGAGAGCCAAGGAGTTGAGTTGTAAAGGCATTGAATGACTTTACAAAGCAATGGCCTGGGGCGGGGTTTGTTTACTTCCCTTTGCCTTACTGACCCCATCTGATCTCAGATAGAAATGGGGTTTTCTGGGAC ...........................................................((((....((((((..............(((((...(((((((....))).)))).......))))).....((((..((((((((.(((...))).))))))))..))))))))))))))...................................................................... ........................................................57..........................................................................................................................181................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532873(GSM532873) G699N. (cervix) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................CTGGGACAGGCCTGGGGGAC......................................................................................................................................................................... | 20 | 37.00 | 0.00 | 12.00 | 7.00 | 3.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | |
| .............................................................CTGGGACAGGCCTGGGGGA.......................................................................................................................................................................... | 19 | 7.00 | 0.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..............................................................TGGGACAGGCCTGGG............................................................................................................................................................................. | 15 | 0 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CTGGGACAGGCCTGGGGGAT......................................................................................................................................................................... | 20 | 2.00 | 0.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................AGCGGACAGATTGCAGTC............................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................TTTACAAAGCAATGGCCTGG......................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CTGGGACAGGCCTGGGGGCC......................................................................................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................AGGCTTGGTGGGTAGAGC.................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CTGGGACAGGCCTGGGGGAG......................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................CAGGCCTGGGAAGTGATAGAGG................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCAGCGGACAGATTGCA.................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TACTCTGGGAGAGAGCCAAGGAGTTGAG................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CTTCCCTTTGCCTTAATTT....................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................CCTGGGACAGGCCTGGGAAGTGATAGAGGC................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCTGGGACAGGCCTGGGGGAC......................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .............................................................CTGGGACAGGCCTGGGGTAC......................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................TCTCAGATAGAAATGGCAG.......... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................GTGGGTAGAGCAATC.............................................................................................................................................. | 15 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGGACAGGCCTGGGAAG.......................................................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ............................................................................................................................................................................CCTGGGGCGGGGTTT............................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| TTTGCCTTTGTGTTTCCAACAGGTCAGCGGACAGATTGCAGTCTAGCCAGGTGAGTGTGGCCTGGGACAGGCCTGGGAAGTGATAGAGGCTTGGTGGGTAGAGCAATCTTACTCTGGGAGAGAGCCAAGGAGTTGAGTTGTAAAGGCATTGAATGACTTTACAAAGCAATGGCCTGGGGCGGGGTTTGTTTACTTCCCTTTGCCTTACTGACCCCATCTGATCTCAGATAGAAATGGGGTTTTCTGGGAC ...........................................................((((....((((((..............(((((...(((((((....))).)))).......))))).....((((..((((((((.(((...))).))))))))..))))))))))))))...................................................................... ........................................................57..........................................................................................................................181................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191598(GSM715708) 79genomic small RNA (size selected RNA from t. (breast) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR189785 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532873(GSM532873) G699N. (cervix) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | DRR001486(DRX001040) "Hela long cytoplasmic cell fraction, LNA(+)". (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................AGATAGAAATGGGGTATA....... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .................AACAGGTCAGCGGACTTT....................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...................AGACTGCAATCTGTCCGCTGACCTG.............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |