| (1) AGO1.ip | (13) BRAIN | (10) BREAST | (15) CELL-LINE | (1) FIBROBLAST | (2) HEART | (5) LIVER | (3) OTHER | (3) SKIN | (2) UTERUS |
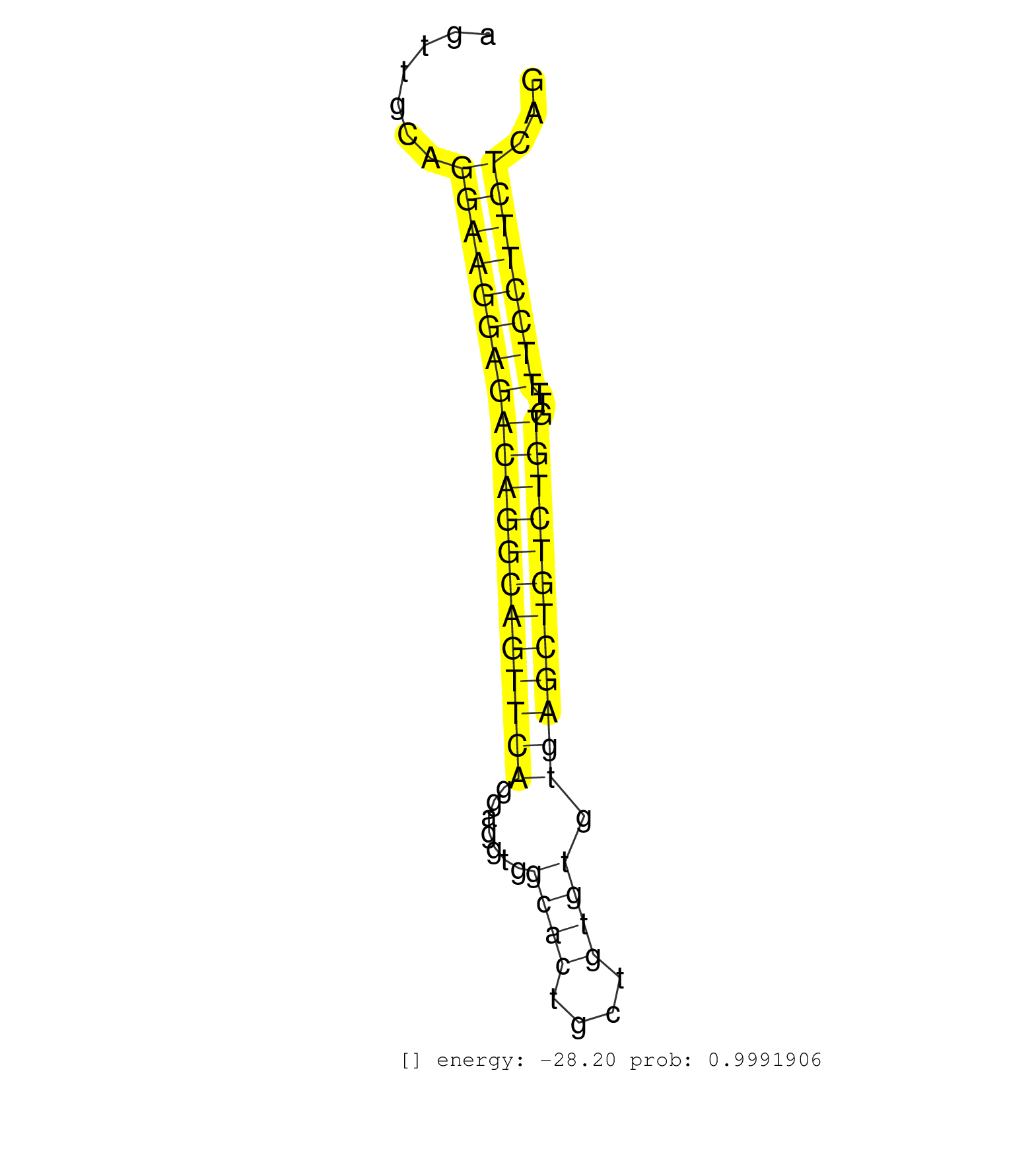
| ATATGATGCAGTGTGAGTTCAATTTCAGTGGAAATGGCAATTTCCATATGGTAAGAAGTGGCAGACCGCGTAATATTAGTAAATGGACATATAGTCAAATCATAACTCATTTCTTTCTATATAGAGAAGCAGTAACCTTAATAATGCAAGTCCATGTTGATCTGAAAGTTCACTTTACATTTCATTTTATAAAGCAATTAGAGATAGAGTCAGACAGTGCAACCCAGAGCCTGGAAAAGATTTTTTTGCT .....................................................................................(((.....)))........((((((((((..((.((((((...(((.............)))..........(((..((((((....)))).))..))).)))))).)).....)))))).))))........................................ ......................................................................71.........................................................................................................................................210...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189785 | SRR029131(GSM416760) MCF7. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577738(Rovira) total RNA. (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577743(Rovira) total RNA. (breast) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577579(Rovira) total RNA. (breast) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191478(GSM715588) 30genomic small RNA (size selected RNA from t. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | TAX577741(Rovira) total RNA. (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR189786 | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577589(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTGA............................................ | 20 | 83.00 | 0.00 | 5.00 | 7.00 | 7.00 | - | 4.00 | 6.00 | 3.00 | 3.00 | 3.00 | 5.00 | 5.00 | 1.00 | 2.00 | 4.00 | 3.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 2.00 | 2.00 | 2.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTG............................................. | 19 | 10.00 | 0.00 | 2.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTTA............................................ | 20 | 7.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACT.............................................. | 18 | 7.00 | 0.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTT............................................. | 19 | 4.00 | 0.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTGT............................................ | 20 | 4.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAATGA............................................ | 20 | 3.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTGC............................................ | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................TATAAAGCAATTAGACTG............................................. | 18 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................TAGAGATAGAGTCAGACAGT................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTAGTAAATGGACATGGAT............................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................ATTTTATAAAGCAATGAGA................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTAA............................................ | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTA............................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGTCTG............................................. | 19 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAATG............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGCCTG............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAATGT............................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................ACATTTCATTTTATAAACCT...................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAAAGA............................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................TCAGACAGTGCAACCCAGAGCC................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CAGTGTGAGTTCAATTTCAGTGGAAATGG..................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TCAATTTCAGTGGAAATGGCAA.................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAAATA............................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................CTTTACATTTCATTTTACG........................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAGTGA............................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGACTCA............................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................AAAGCAATTAGAGATCACA......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .......................................................................................................................TATAGAGAAGCAGTACCGT................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATAAAGCAATTAGAACGA............................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................TAGAGAAGCAGTAAC.................................................................................................................. | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATATGATGCAGTGTGAGTTCAATTTCAGTGGAAATGGCAATTTCCATATGGTAAGAAGTGGCAGACCGCGTAATATTAGTAAATGGACATATAGTCAAATCATAACTCATTTCTTTCTATATAGAGAAGCAGTAACCTTAATAATGCAAGTCCATGTTGATCTGAAAGTTCACTTTACATTTCATTTTATAAAGCAATTAGAGATAGAGTCAGACAGTGCAACCCAGAGCCTGGAAAAGATTTTTTTGCT .....................................................................................(((.....)))........((((((((((..((.((((((...(((.............)))..........(((..((((((....)))).))..))).)))))).)).....)))))).))))........................................ ......................................................................71.........................................................................................................................................210...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189785 | SRR029131(GSM416760) MCF7. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577738(Rovira) total RNA. (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577743(Rovira) total RNA. (breast) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577579(Rovira) total RNA. (breast) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191478(GSM715588) 30genomic small RNA (size selected RNA from t. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | TAX577741(Rovira) total RNA. (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR189786 | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | TAX577589(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................AGACAGTGCAACCCAGTGA.................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................CAAGTCCATGTTGATGTG...................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ..........................................................................................................................................................................................................................GCAACCCAGAGCCTGTTGG............. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................AGTTCACTTTACATTCTG.................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............GAGTTCAATTTCAGTGCT.......................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................AGAAGCAGTAACCTTGGC............................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |