| (5) B-CELL | (2) BRAIN | (5) BREAST | (3) CELL-LINE | (4) CERVIX | (5) HEART | (1) KIDNEY | (2) LIVER | (1) OTHER | (9) SKIN | (1) TESTES | (1) UTERUS | (1) XRN.ip |
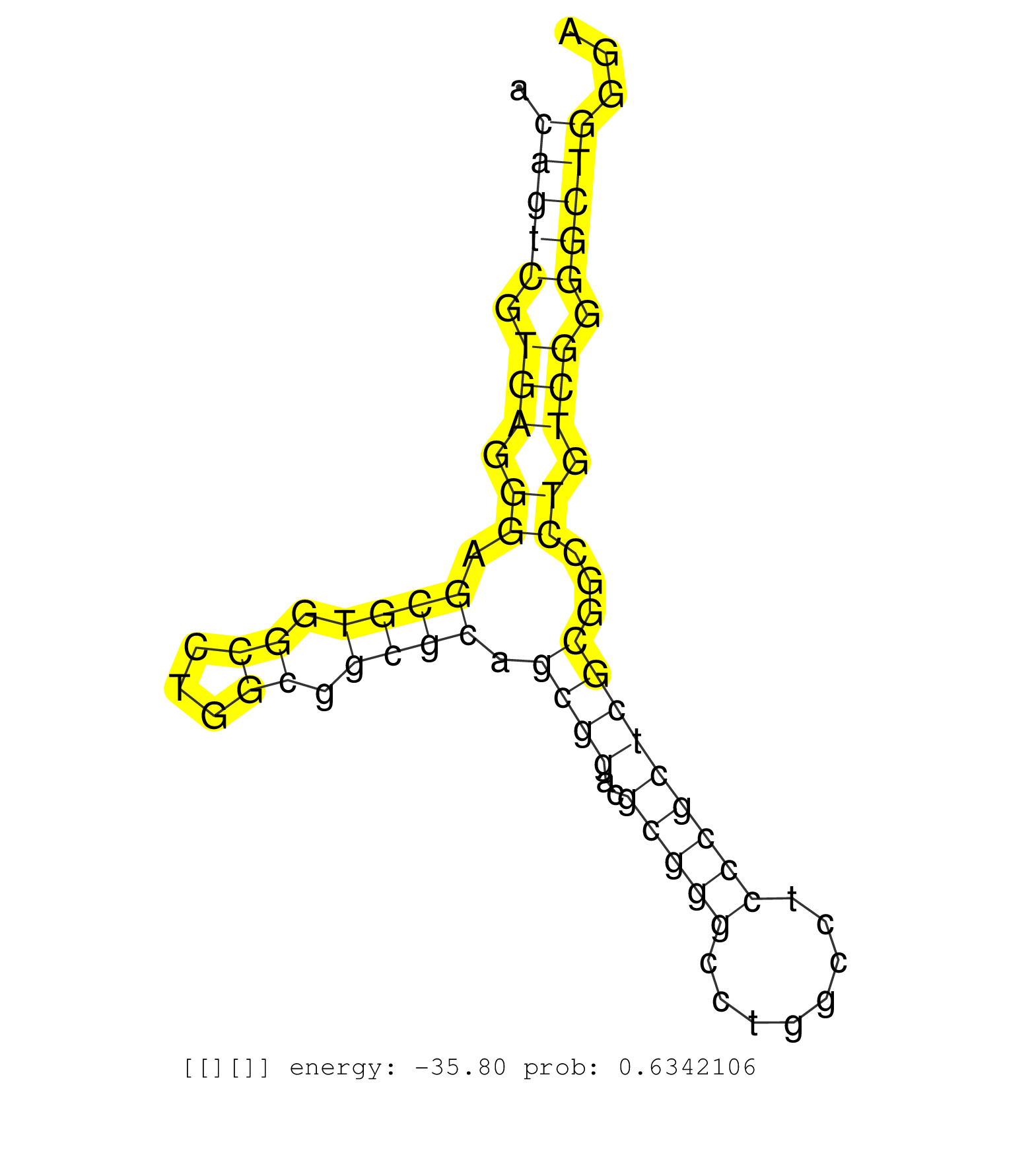
| GTCTAAAATGACCCACAATGCTTTGGAGAACATTAACGTGATTGGCCAAGGCTTGAAGCATCTCTTCCAGCACCAGCGCAGGAGGTCATCAGTGTCTCCACATGATGTGCAGCAAATTCAGGCAGATCCAGAACCTGAAATGGATCTGGAAAGCCAGAACGCATGTGCTGAGATTGATGGTGTCCCCACCCACCCCACAGCTCTGAATCGTGTCCTGCAGCAGATTCGAGTGCCACCCAAGATGAAGAGA ..........................................................((((...((((((((..((((((((((.((....)))))))......))))).((...(((.(((...((((((.((......)).))))))..))).))).))..)))))).))..))))....................................................................... ..................................................51...............................................................................................................................180.................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR040037(GSM532922) G243T. (cervix) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR040035(GSM532920) G001T. (cervix) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577745(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR040028(GSM532913) G026N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GCTTGAAGCATCTCTGGAC..................................................................................................................................................................................... | 19 | 27.00 | 0.00 | 15.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GCTTGAAGCATCTCTGG....................................................................................................................................................................................... | 17 | 9.00 | 0.00 | 4.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GCTTGAAGCATCTCTGGA...................................................................................................................................................................................... | 18 | 3.00 | 0.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GCTTGAAGCATCTCTGTAC..................................................................................................................................................................................... | 19 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................CATGTGCTGAGATTGATGGTGTCCCCAC............................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CTGAGATTGATGGTGTCCCCACCCACCCCAC.................................................... | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CCAGAACGCATGTGCTGAGATTGATG....................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TAACGTGATTGGCCAAGGCTTGAAGC............................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CATTAACGTGATTGGCCAAGGC...................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GCTGAGATTGATGGTGTCCCCACCCACCCCAC.................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTAACGTGATTGGCCAAGGCTTGAAGC............................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCTTTGGAGAACATTAACGTGATTGGCC........................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TCCTGCAGCAGATTCGAGTGT................. | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................CGCATGTGCTGAGATTGATGGTGTCCCCA.............................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................AGAACGCATGTGCTGAGATTG.......................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCTTGAAGCATCTCTGT....................................................................................................................................................................................... | 17 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................GAAATGGATCTGGAATTT................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................TCCACATGATGTGCAGCAAATTCAGGCA.............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGCTTTGGAGAACATTAAC..................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CCTGCAGCAGATTCGAG.................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................AACGTGATTGGCCAAGGCTTGAAGCATCTC.......................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TCTGGAAAGCCAGAACGCA....................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CACCAGCGCAGGAGGTC................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................AACATTAACGTGATTGGCCAAGGCTTGAAG................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCTTGAAGCATCTCTGGAA..................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................ATGCTTTGGAGAACATTAACA.................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................ACCCCACAGCTCTGAATCGTGTCCTGCAGC............................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CACAGCTCTGAATCGTGTCCTGCAGC............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ACGTGATTGGCCAAGGCTTGAAGC............................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCTTGAAGCATCTCTTGATT.................................................................................................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................TTAACGTGATTGGCCAAGGCTTGAAGCA.............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AATCGTGTCCTGCAGC............................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GCTTGAAGCATCTCTTGA...................................................................................................................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................AGAACGCATGTGCTGAGATTGAT........................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TAACGTGATTGGCCAAGGCTTGAAGCA.............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CATGTGCTGAGATTGATGGTGTC.................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................CTGAATCGTGTCCTGCAGCA............................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............ACAATGCTTTGGAGAACATTAACGTGATTGGC............................................................................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ATCGTGTCCTGCAGCAGAT......................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCTTGAAGCATCTCTGGC...................................................................................................................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................ATCTCTTCCAGCACCTAG............................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........TGACCCACAATGCTTTGGAGAACATTAACGTGT................................................................................................................................................................................................................. | 33 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................AGGCAGATCCAGAACCTG................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..................................................................................................................................GAACCTGAAATGGATCTGGAAAGCC............................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GAAATGGATCTGGAAAGC................................................................................................ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| .......................................................................................................................................TGAAATGGATCTGGAAAGC................................................................................................ | 19 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| .......................................................................................................................................TGAAATGGATCTGGAAAG................................................................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ......................................................................................................................................CTGAAATGGATCTGGA.................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGAAATGGATCTGGAA................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GTCTAAAATGACCCACAATGCTTTGGAGAACATTAACGTGATTGGCCAAGGCTTGAAGCATCTCTTCCAGCACCAGCGCAGGAGGTCATCAGTGTCTCCACATGATGTGCAGCAAATTCAGGCAGATCCAGAACCTGAAATGGATCTGGAAAGCCAGAACGCATGTGCTGAGATTGATGGTGTCCCCACCCACCCCACAGCTCTGAATCGTGTCCTGCAGCAGATTCGAGTGCCACCCAAGATGAAGAGA ..........................................................((((...((((((((..((((((((((.((....)))))))......))))).((...(((.(((...((((((.((......)).))))))..))).))).))..)))))).))..))))....................................................................... ..................................................51...............................................................................................................................180.................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040007(GSM532892) G601T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR040037(GSM532922) G243T. (cervix) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR040035(GSM532920) G001T. (cervix) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577745(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR040028(GSM532913) G026N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......AATGACCCACAATGCGGT.................................................................................................................................................................................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................ACGATTCAGAGCTGTGGGG....................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CATGATGTGCAGCAAGTTT................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................CTGCTGCAGGACACGATTCAG........................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................ATCTGCTGCAGGACACGATT......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CATCTCTTCCAGCACCGTA............................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................GCAGGACACGATTCAGAG................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................CTGCTGCAGGACACGATTCAGAG........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CATCTCTTCCAGCACCAACA............................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......AATGACCCACAATGCGGTT................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ......AATGACCCACAATGCGGTA................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................GGGTGGGGACACCATC........................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |