| (1) AGO2.ip | (6) B-CELL | (4) BRAIN | (13) BREAST | (6) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (2) HEART | (1) LIVER | (1) OTHER | (1) SKIN | (4) UTERUS | (1) XRN.ip |
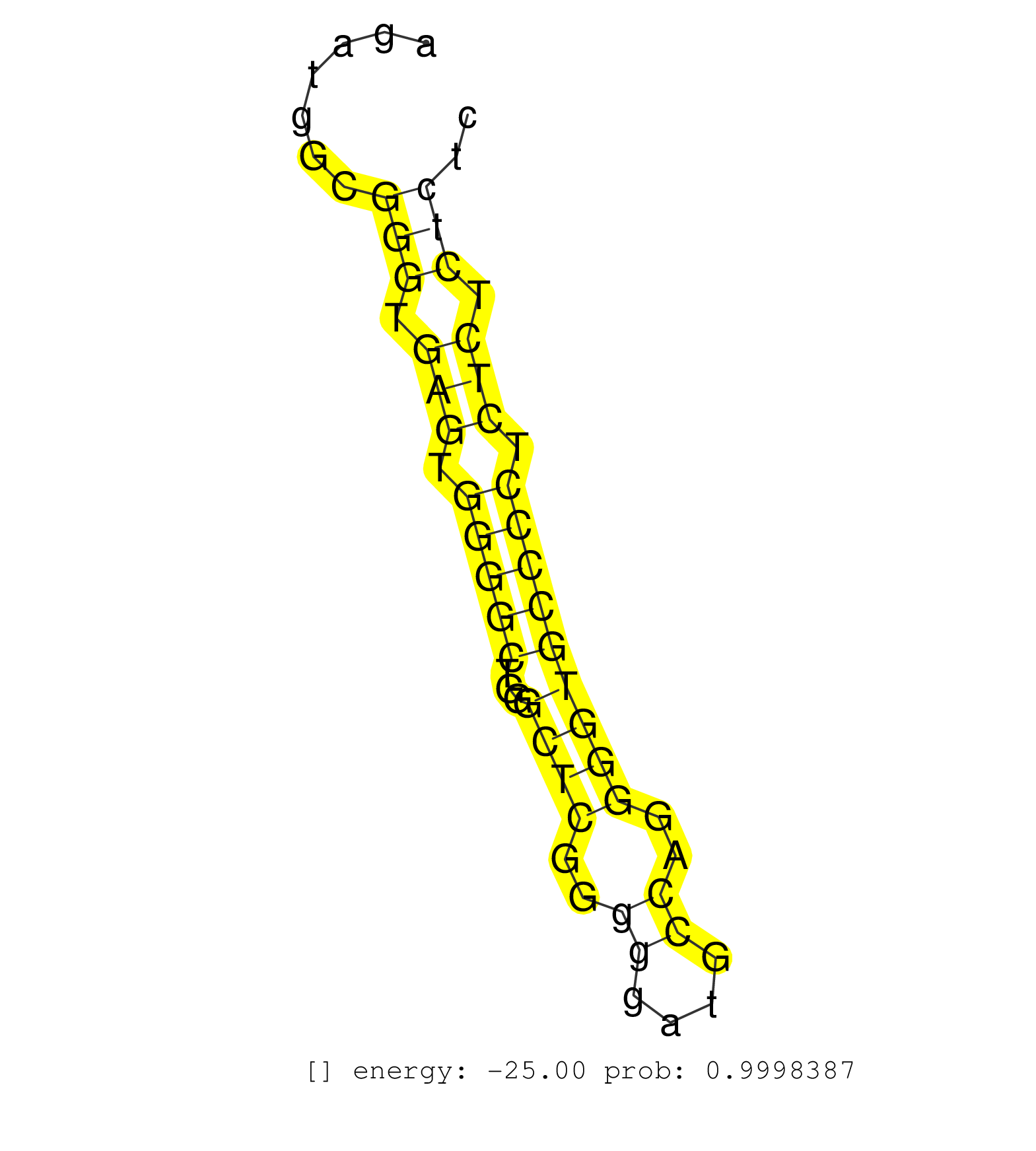
| CCACTCACTCGCCTGGAGAAGTATGCTTTCAGCGAGAACACCTTCAACCGGTAAGCCCAACCAGGCCTGTGTATGTTGACAACTCTGTGTTGTGTGCTTTAAGCTGACACTTGGGGCCACTTCTCTGTGACAGCTTTGAGGAGACTTCATGGATGATGAGGTAGGTTGGATACAGATTGTCACCAGGATGCTGGGCTTCTGTCTGAATGGTACCATCTTATTAGGCTTTTTCATTATTTTTGTTATTGTT ...............................................................(((((((((......(((((.....)))))..((.....))..))))...))))).....((((((.((((((.......(((((((.....)))))))))))))..)))))).......................................................................... ...............................................................64.................................................................................................................179..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189787 | SRR189786 | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189784 | TAX577580(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | TAX577742(Rovira) total RNA. (breast) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189782 | SRR040008(GSM532893) G727N. (cervix) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191445(GSM715555) 110genomic small RNA (size selected RNA from . (breast) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189785 | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................TGAGGTAGGTTGGATAGTT........................................................................... | 19 | 32.00 | 0.00 | 6.00 | 5.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | 2.00 | 1.00 | 2.00 | - | 2.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCGG................................................................................................................................................................... | 19 | 1 | 7.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................GTGTATGTTGACAACTCGGTA................................................................................................................................................................. | 21 | 1 | 5.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGT............................................................................ | 18 | 4.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCGGT.................................................................................................................................................................. | 20 | 1 | 3.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGGTAGGTTGGATGGTT........................................................................... | 19 | 2.00 | 0.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATAG............................................................................. | 17 | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGTTT.......................................................................... | 20 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGTC........................................................................... | 19 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGCT........................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATTGGT........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................................TGATGAGGTAGGTTGTGTG.............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCG.................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CATCTTATTAGGCTTATGC.................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATTTTT........................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCGA................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGGTAGGTTGGATGGT............................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCGTTA................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGTTA.......................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................TGATGAGGTAGGTTGATTG.............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTC..................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGGTAGGTTGGATGGCT........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATTGTT........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GTGTATGTTGACAACTCTT................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................TGATGAGGTAGGTTGTA................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATAGGT........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TGAGGTAGGTTGGATTTTG........................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................GCTGACACTTGGGGCCACTTCTCTG........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................GTGTATGTTGACAACTCGTT.................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TTTCATTATTTTTGTTATT... | 19 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| CCACTCACTCGCCTGGAGAAGTATGCTTTCAGCGAGAACACCTTCAACCGGTAAGCCCAACCAGGCCTGTGTATGTTGACAACTCTGTGTTGTGTGCTTTAAGCTGACACTTGGGGCCACTTCTCTGTGACAGCTTTGAGGAGACTTCATGGATGATGAGGTAGGTTGGATACAGATTGTCACCAGGATGCTGGGCTTCTGTCTGAATGGTACCATCTTATTAGGCTTTTTCATTATTTTTGTTATTGTT ...............................................................(((((((((......(((((.....)))))..((.....))..))))...))))).....((((((.((((((.......(((((((.....)))))))))))))..)))))).......................................................................... ...............................................................64.................................................................................................................179..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189787 | SRR189786 | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189784 | TAX577580(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | TAX577742(Rovira) total RNA. (breast) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343334 | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR189782 | SRR040008(GSM532893) G727N. (cervix) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191445(GSM715555) 110genomic small RNA (size selected RNA from . (breast) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189785 | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................GTGACAGCTTTGAGGAG........................................................................................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ....................................................................................CTGTGTTGTGTGCTTTACCTT................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................GCCCAACCAGGCCTGGGT.................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................GATGAGGTAGGTTGGTAAG............................................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................ACACCTTCAACCGGTGTT................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .......................................................................................................................................................GATGATGAGGTAGGTTGTAAA.............................................................................. | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................CAACCTACCTCATCAT.................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ........................................................................................................................................................ATGATGAGGTAGGTTGG................................................................................. | 17 | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | |
| ...............................................................................................................................................................TGTATCCAACCTACC............................................................................ | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |