| (1) AGO1.ip | (10) BRAIN | (2) BREAST | (1) CELL-LINE | (1) FIBROBLAST | (1) LIVER | (3) OTHER | (3) SKIN | (2) UTERUS |
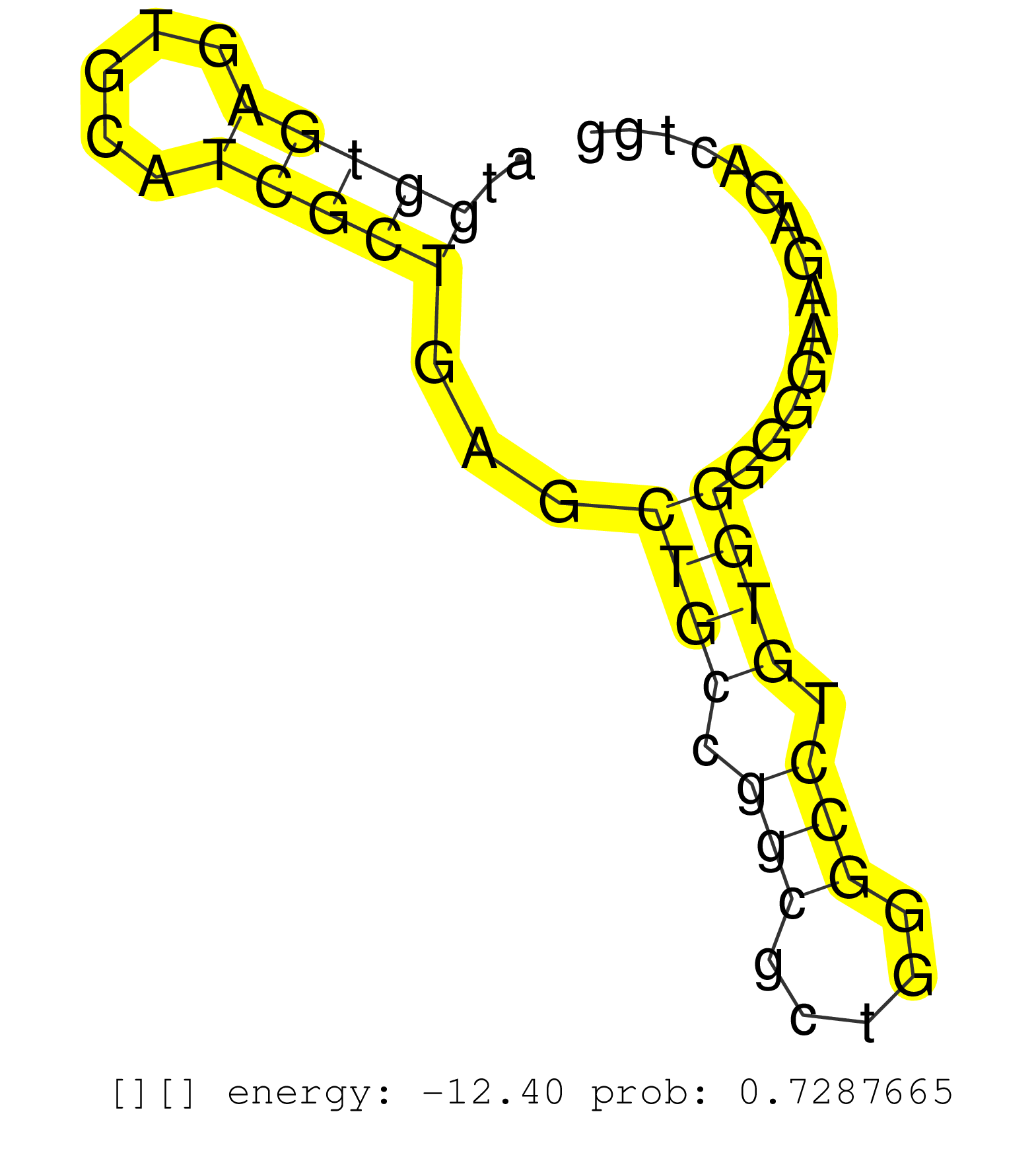
| TCGCAAGTCCTCTGCAAGGATTTACGCTTCTGCCTTCTCTACACTGCCAGGTCAGCTACTATTCCTCCAAACTGCTTCACATCTTCCAAAAATATGTCATTCTCTCATGTTGACTGCTGTCTCTCTCTTGGCTCTTTGTGTATGTGTATGTATAGTTGTGTGTTGCGGGGGTAGGGTGAGATAGAACAGGAACCCCCTCTTAGGGACCTGGATGCCCCACCAAACATAAATGTTTTTTATATTTTAAATT ....................................................................................................................(((((((.((((..((.(((((((.(((..((........))..)))))))))))))))).))))))).................................................................. ...............................................................................................................112........................................................................187............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577580(Rovira) total RNA. (breast) | SRR189784 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343334 | SRR444045(SRX128893) Sample 6cDNABarcode: AF-PP-341: ACG CTC TTC C. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | TAX577740(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAACAG................................................................... | 20 | 78.00 | 0.00 | 72.00 | 2.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAATAG................................................................... | 20 | 55.00 | 0.00 | 47.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGTA...................................................................... | 17 | 7.00 | 0.00 | - | 1.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGTAT..................................................................... | 18 | 3.00 | 0.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAA...................................................................... | 17 | 3.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAAGAG................................................................... | 20 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAACA.................................................................... | 19 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAATA.................................................................... | 19 | 2.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAACAA................................................................... | 20 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGTATA.................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGTATT.................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................CTGTCTCTCTCTTGGCTCTTTGT............................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAGAGGTG................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAAT..................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAGCAGC.................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAACGG................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAGCGGA.................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGAGATAGAACAGGACAG........................................................ | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAAAAG................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TGCGGGGGTAGGGTGAATAA................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................CCTTCTCTACACTGCCA......................................................................................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCGCAAGTCCTCTGCAAGGATTTACGCTTCTGCCTTCTCTACACTGCCAGGTCAGCTACTATTCCTCCAAACTGCTTCACATCTTCCAAAAATATGTCATTCTCTCATGTTGACTGCTGTCTCTCTCTTGGCTCTTTGTGTATGTGTATGTATAGTTGTGTGTTGCGGGGGTAGGGTGAGATAGAACAGGAACCCCCTCTTAGGGACCTGGATGCCCCACCAAACATAAATGTTTTTTATATTTTAAATT ....................................................................................................................(((((((.((((..((.(((((((.(((..((........))..)))))))))))))))).))))))).................................................................. ...............................................................................................................112........................................................................187............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577580(Rovira) total RNA. (breast) | SRR189784 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343334 | SRR444045(SRX128893) Sample 6cDNABarcode: AF-PP-341: ACG CTC TTC C. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | TAX577740(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................TTTTTATATTTTAAATCAAG | 20 | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGTGTATGTATAGTTGTGCTC....................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................GGGGGTAGGGTGAGACC................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................CTGCTGTCTCTCTCTTAT....................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................CTGCTGTCTCTCTCTTCT....................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................TTCACATCTTCCAAAAATTGTC......................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |