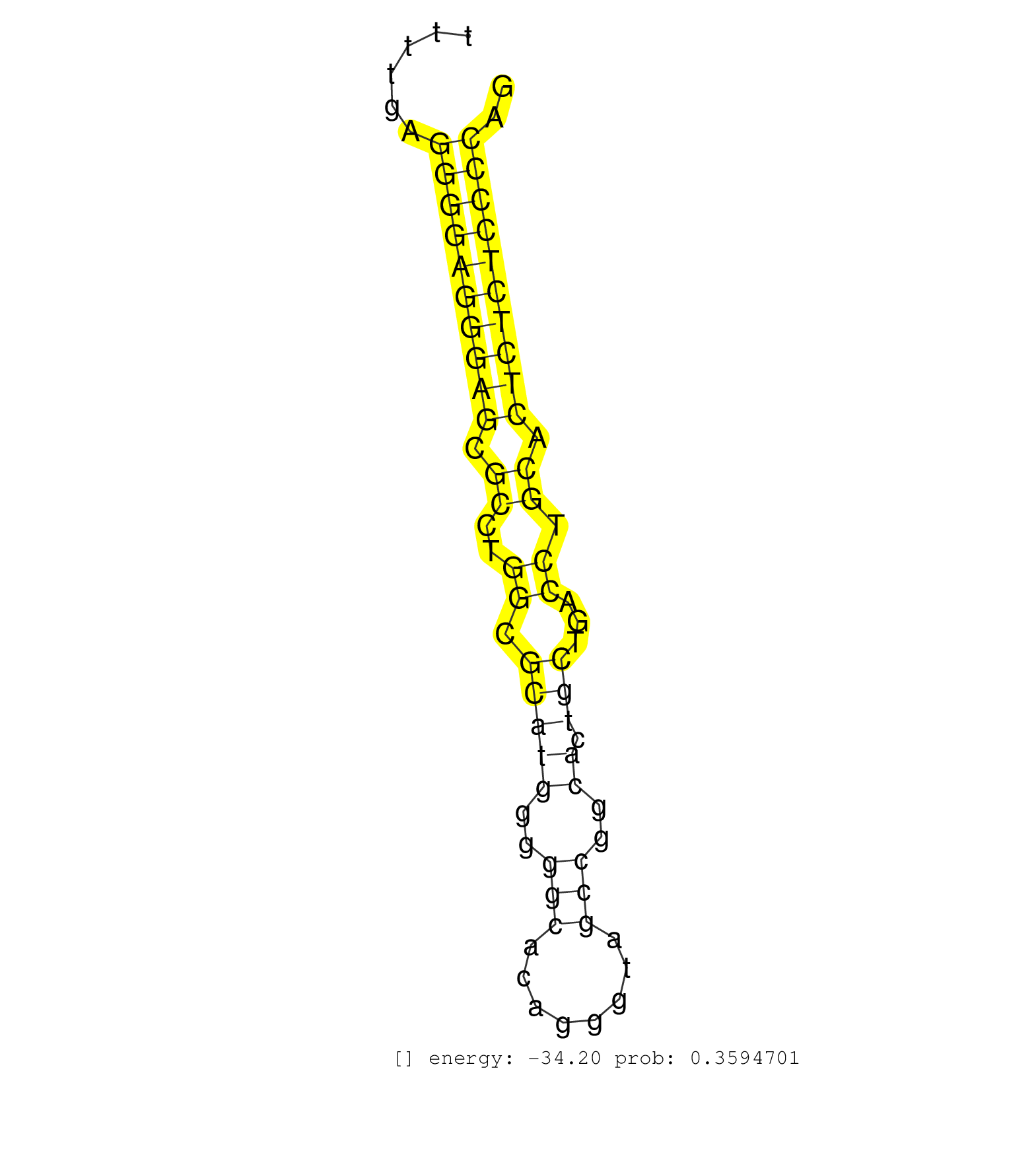
| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (3) AGO2.ip | (1) AGO3.ip | (3) B-CELL | (6) BRAIN | (6) BREAST | (30) CELL-LINE | (1) CERVIX | (8) HEART | (3) HELA | (1) KIDNEY | (5) LIVER | (2) OTHER | (2) RRP40.ip | (33) SKIN | (1) TESTES | (2) XRN.ip |
| ACCGGTTTATCGAGAAACCCGTGGTTTACTGGCGAGGGAAGCAGCAGCGGGTTACGGCACCAAATGCTGGGGCGGTGGCGGGTGGGAAGCAGTGCCAAGGACAATTCCTGCTCTCCCAACATAGGCTTTTGAGGGGAGGGAGCGCCTGGCGCATGGGGGCACAGGGTAGCCGGCACTGCTGACCTGCACTCTCTCCCCAGGAGCACAGAAAATGGTTCTGCCCCCTCCACAGCTCCCACAGACCAGAGCC ..................................................................((((((.((..((((((....(((((((((..((.....((((((((((((..((........))..))))))).((.((((.....)))).)).)))))...))))))))))).))))))....)).)))))).................................................. ..................................................................67...................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR189783 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189784 | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189787 | SRR189785 | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR037938(GSM510476) 293Red. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR191451(GSM715561) 177genomic small RNA (size selected RNA from . (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR191599(GSM715709) 89genomic small RNA (size selected RNA from t. (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029128(GSM416757) H520. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191531(GSM715641) 134genomic small RNA (size selected RNA from . (breast) | SRR040014(GSM532899) G623N. (cervix) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGT................................................. | 23 | 1 | 136.00 | 55.00 | 51.00 | 11.00 | 14.00 | 4.00 | 2.00 | 8.00 | 5.00 | 1.00 | 1.00 | - | - | 4.00 | - | - | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAG.................................................. | 22 | 1 | 55.00 | 55.00 | 3.00 | 28.00 | 3.00 | 7.00 | 1.00 | - | - | - | - | 4.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGA................................................. | 23 | 1 | 43.00 | 55.00 | 16.00 | - | 13.00 | - | 1.00 | 3.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCAGT................................................. | 22 | 1 | 18.00 | 1.00 | 5.00 | 4.00 | 5.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCA................................................... | 21 | 1 | 16.00 | 16.00 | 4.00 | 6.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGTA................................................ | 24 | 1 | 7.00 | 55.00 | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCAGA................................................. | 22 | 1 | 6.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAA.................................................. | 22 | 1 | 6.00 | 16.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAAAT................................................ | 24 | 1 | 4.00 | 16.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGTT................................................ | 24 | 1 | 4.00 | 55.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACTGCTGACCTGCACTCTCTCCCCAGA................................................. | 27 | 1 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAAT................................................. | 23 | 1 | 3.00 | 16.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAT.................................................. | 22 | 1 | 3.00 | 16.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCC.................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................ACTGCTGACCTGCACTCTCTCCCCAGAA................................................ | 28 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCCGT................................................. | 23 | 1 | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CTGCTGACCTGCACTCTCTCCCCAG.................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CACTGCTGACCTGCACTCTCTCCCCAGA................................................. | 28 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCC.................................................... | 19 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AGAAAATGGTTCTGCCCCCTCCACA................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGAA................................................ | 24 | 1 | 2.00 | 55.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GACCTGCACTCTCTCCCCAGTT................................................ | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................AGGGGAGGGAGCGCCTGGCGCATGGGGGCA......................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTTACGGCACCA............................................................................................................................................................................................ | 15 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAATA................................................ | 24 | 1 | 2.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGAT................................................ | 24 | 1 | 2.00 | 55.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................ACCTGCACTCTCTCCCCAGA................................................. | 20 | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGC................................................. | 23 | 1 | 2.00 | 55.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCCGG................................................. | 22 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGACT............................................... | 25 | 1 | 1.00 | 55.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCAC.................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................GCGGTGGCGGGTGGGCC.................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCTG.................................................. | 22 | 1 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AGGGGAGGGAGCGCCTGGAAT.................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................TCCCCAGGAGCACAGACTTG..................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................CGGCACTGCTGACCTGCACTCTCTCCCCA................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCAGTTT............................................... | 25 | 1 | 1.00 | 55.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................GCGGGTTACGGCACCAAATG........................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................CCCCAGGAGCACAGAAAATG.................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCATT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GCTGACCTGCACTCTCTCCCCAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GGCGGTGGCGGGTGGTGC.................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACTGCTGACCTGCACTCTCTCCCCAG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGGGGCGGTGGCGGGTGTG.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................ACTGCTGACCTGCACTCTCTCCCCAGAAA............................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AGGGGAGGGAGCGCCTGGCGCAA................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CGCATGGGGGCACAGCGGG.................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................AGGGTAGCCGGCACTGCTGACCGCAC.............................................................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................CCAGGAGCACAGAAAAT..................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCCGA.................................................. | 22 | 1 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCTCAG.................................................. | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................CACTGCTGACCTGCACTCTCTCCCC.................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................CCCCAGGAGCACAGAAAA...................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCTCCCAACATAGGCTTTTGAGGGGAGGG.............................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTGCACTCTCTCCCTAG.................................................. | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGACCTGCACTCTCTCCCCAGAA................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AGGGGAGGGAGCGCCTGGCGCA................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AGAAAATGGTTCTGCCCCCTC....................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCTGCACTCTCTCCCCAGT................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................AGGGGAGGGAGCGCCTG...................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ACCGGTTTATCGAGAAACCCGTGGTTTACTGGCGAGGGAAGCAGCAGCGGGTTACGGCACCAAATGCTGGGGCGGTGGCGGGTGGGAAGCAGTGCCAAGGACAATTCCTGCTCTCCCAACATAGGCTTTTGAGGGGAGGGAGCGCCTGGCGCATGGGGGCACAGGGTAGCCGGCACTGCTGACCTGCACTCTCTCCCCAGGAGCACAGAAAATGGTTCTGCCCCCTCCACAGCTCCCACAGACCAGAGCC ..................................................................((((((.((..((((((....(((((((((..((.....((((((((((((..((........))..))))))).((.((((.....)))).)).)))))...))))))))))).))))))....)).)))))).................................................. ..................................................................67...................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR189783 | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189784 | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189787 | SRR189785 | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR037938(GSM510476) 293Red. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR191451(GSM715561) 177genomic small RNA (size selected RNA from . (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR191599(GSM715709) 89genomic small RNA (size selected RNA from t. (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029128(GSM416757) H520. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191531(GSM715641) 134genomic small RNA (size selected RNA from . (breast) | SRR040014(GSM532899) G623N. (cervix) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR029130(GSM416759) DLD2. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TCCTGCTCTCCCAACCTGT.............................................................................................................................. | 19 | 53.00 | 0.00 | - | - | - | - | 6.00 | - | - | 4.00 | 6.00 | - | - | - | 4.00 | - | - | 1.00 | 1.00 | 3.00 | - | - | - | - | 3.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | 2.00 | - | 1.00 | 2.00 | - | 2.00 | - | - | - | - | 2.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ............................................................................................................TGCTCTCCCAACATATTA............................................................................................................................ | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................GGGGCGGTGGCGGGTCTCT................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................GCCCCCTCCACAGCTCCCC............ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................TCCTGCTCTCCCAACCTGC.............................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................CCCCTCCACAGCTCCCAT........... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................AATGGTTCTGCCCCCTCGGC.................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ......................................................................................................AATTCCTGCTCTCCCATTGG................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................ATTCCTGCTCTCCCAAATG................................................................................................................................ | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................AAGGACAATTCCTGCTCG........................................................................................................................................ | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................GTCTGTGGGAGCTGT....... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |