| (2) AGO1.ip | (2) AGO2.ip | (2) AGO3.ip | (12) B-CELL | (1) BRAIN | (3) BREAST | (23) CELL-LINE | (1) CERVIX | (4) HELA | (2) LIVER | (1) OTHER | (3) SKIN | (1) TESTES | (1) XRN.ip |
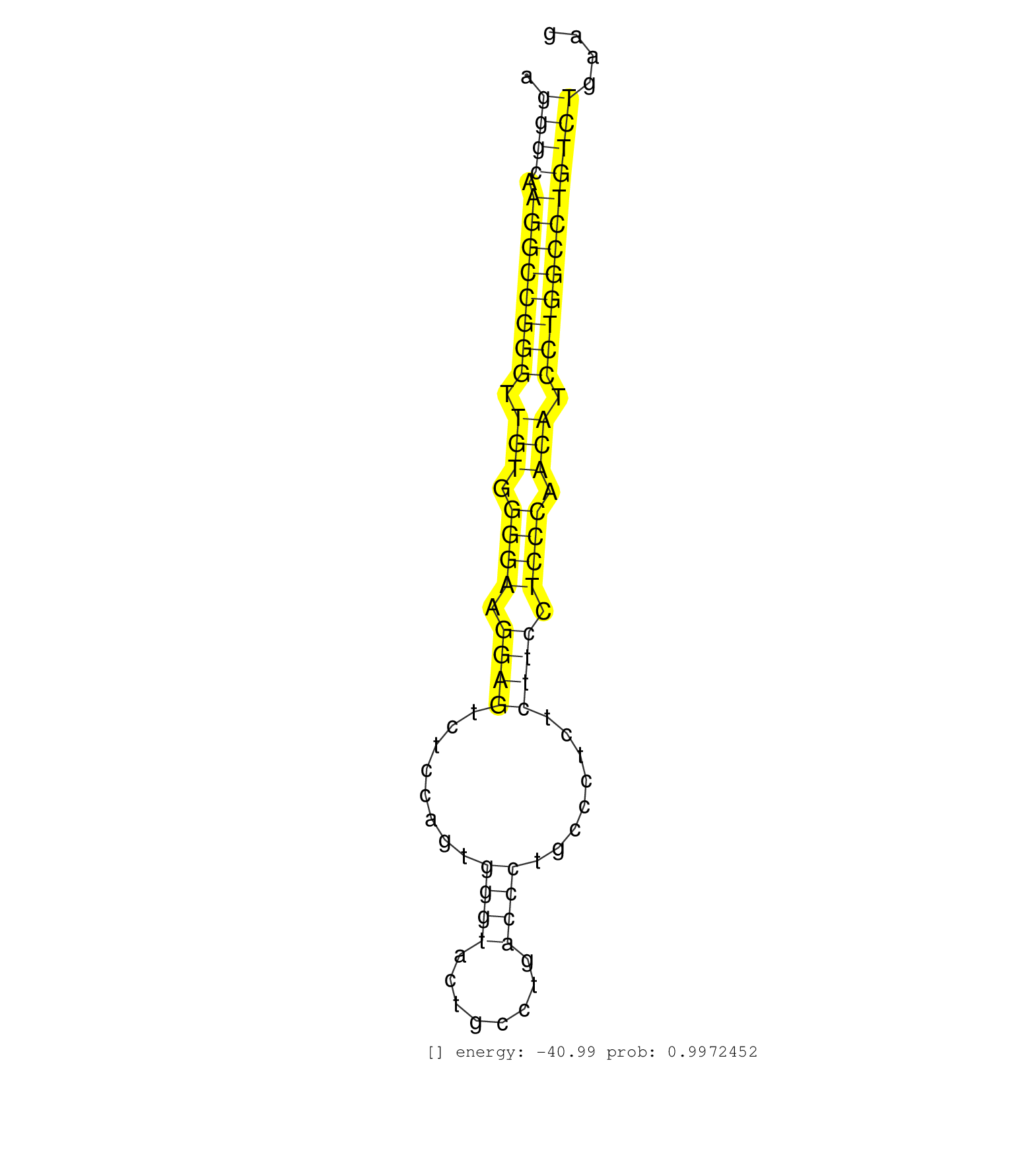
| GCCACGTGGTGCTCAGCACCTTGTGGGAGCCGCATATCACCCAACGCCAGGTTGGTGCCCAGGGCAAGGCCGGGTTGTGGGGAAGGAGTCTCCAGTGGGTACTGCCTGACCCTGCCCTCTCTTCCTCCCAACATCCTGGCCTGTCTGAAGGTGGAGAAAGCCTGGGCCCTCTTGCAGAAGCGCATTCCCAAGACTCATCA .............................................................((((.((((((((.(((.((((.((((........((((........))))........)))).)))).))).))))))))))))...................................................... ............................................................61.......................................................................................150................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR189784 | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001482(DRX001036) "Hela long total cell fraction, control". (hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR189787 | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040036(GSM532921) G243N. (cervix) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR189785 | SRR038858(GSM458541) MEL202. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR191411(GSM715521) 21genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................AAGGCCGGGTTGTGGGGAAGGAT................................................................................................................ | 23 | 1 | 10.00 | 10.00 | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - |
| .................................................................AAGGCCGGGTTGTGGGGAAGGA................................................................................................................. | 22 | 1 | 10.00 | 10.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGCCTGGGCCCTCTTGCAGAAGCGC................. | 25 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGAAGGA................................................................................................................. | 23 | 1 | 7.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGAAG................................................................................................................... | 21 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | - | - | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAAGGAA................................................................................................................ | 23 | 1 | 6.00 | 10.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAAGGATT............................................................................................................... | 24 | 1 | 4.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGAAGGT................................................................................................................. | 23 | 1 | 3.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACTGCCTGACCCTGCCGGG................................................................................. | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................AAGCCTGGGCCCTCTTGCAGA...................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................CTCCCAACATCCTGGCCTGTCT...................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGAAGG.................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGGCAAGGCCGGGTTGTGGGGATA................................................................................................................... | 25 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................TCACCCAACGCCAGGAGC.................................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................GAAAGCCTGGGCCCTCTTGCAGAAGCGCATTC............. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................AAAGCCTGGGCCCTCTTGCAGAA..................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CAACATCCTGGCCTGTCTGAAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GCCTGGGCCCTCTTGCGGCT..................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................GTTGTGGGGAAGGAGCTAC............................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................CAAGGCCGGGTTGTGGGGAAT................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................AGAAGCGCATTCCCAAGACTCAT.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGACTGA................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GCGCATTCCCAAGACTCATCA | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAA.................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................AGGCCGGGTTGTGGGGAAGGAT................................................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ................................................................CAAGGCCGGGTTGTGGGGAAGGTT................................................................................................................ | 24 | 1 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGG........................................................................................................................ | 15 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAAGGATAAA............................................................................................................. | 26 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CTGCCTGACCCTGCCGG.................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................CTGGGCCCTCTTGCAGAAGCG.................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAAG................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TCTTGCAGAAGCGCATTCCC........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAGGCCGGGTTGTGGGGAAGGAAA............................................................................................................... | 24 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGGCCGGGTTGTGGGGAAGGATA............................................................................................................... | 25 | 1 | 1.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTGGTGCCCAGGGCAAGGCCGGGTTGTGGGGAAGGAGTCTCCAGTGGGTACTGCCTGACCCTGCCCTCTCTTCCTCCCAACATCCTGGCCTGTCTGAAG.................................................. | 100 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TACTGCCTGACCCTGCCGGG................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................CAAGGCCGGGTTGTGGGGATGGA................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................GAAAGCCTGGGCCCTCTTGCAGAT..................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................CTCCCAACATCCTGGCCTGT........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ACCTTGTGGGAGCCGCATATA.................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................AGGCCGGGTTGTGGGGAAGGAGTCTC............................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CCTGGGCCCTCTTGCAGAAGCGCAT............... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CCTCCCAACATCCTGGCCTGT........................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................CCAACATCCTGGCCTGTCTGAT................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........GTGCTCAGCACCTTGTGGGAGCCGCA...................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCACGTGGTGCTCAGCACCTTGTGGGAGCCGCATATCACCCAACGCCAGGTTGGTGCCCAGGGCAAGGCCGGGTTGTGGGGAAGGAGTCTCCAGTGGGTACTGCCTGACCCTGCCCTCTCTTCCTCCCAACATCCTGGCCTGTCTGAAGGTGGAGAAAGCCTGGGCCCTCTTGCAGAAGCGCATTCCCAAGACTCATCA .............................................................((((.((((((((.(((.((((.((((........((((........))))........)))).)))).))).))))))))))))...................................................... ............................................................61.......................................................................................150................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR189784 | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR037935(GSM510473) 293cand3. (cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR038852(GSM458535) QF1160MB. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001482(DRX001036) "Hela long total cell fraction, control". (hela) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR189787 | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR343332(GSM796035) "KSHV (HHV8), EBV (HHV-4)". (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040036(GSM532921) G243N. (cervix) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR189785 | SRR038858(GSM458541) MEL202. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR191411(GSM715521) 21genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR038863(GSM458546) MM603. (cell line) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................TCACCCAACGCCAGGTGCAG................................................................................................................................................ | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CCTCCCAACATCCTGGCCTTGA....................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................AGGCCAGGATGTTGGGAGGA.......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGCCGGGTTGTGGGGAATGA................................................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................AACATCCTGGCCTGTTGTG.................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................TGGGAGGAAGAGAGGGC...................................................................... | 17 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |