| (5) CELL-LINE | (1) HEART | (2) OTHER | (2) SKIN |
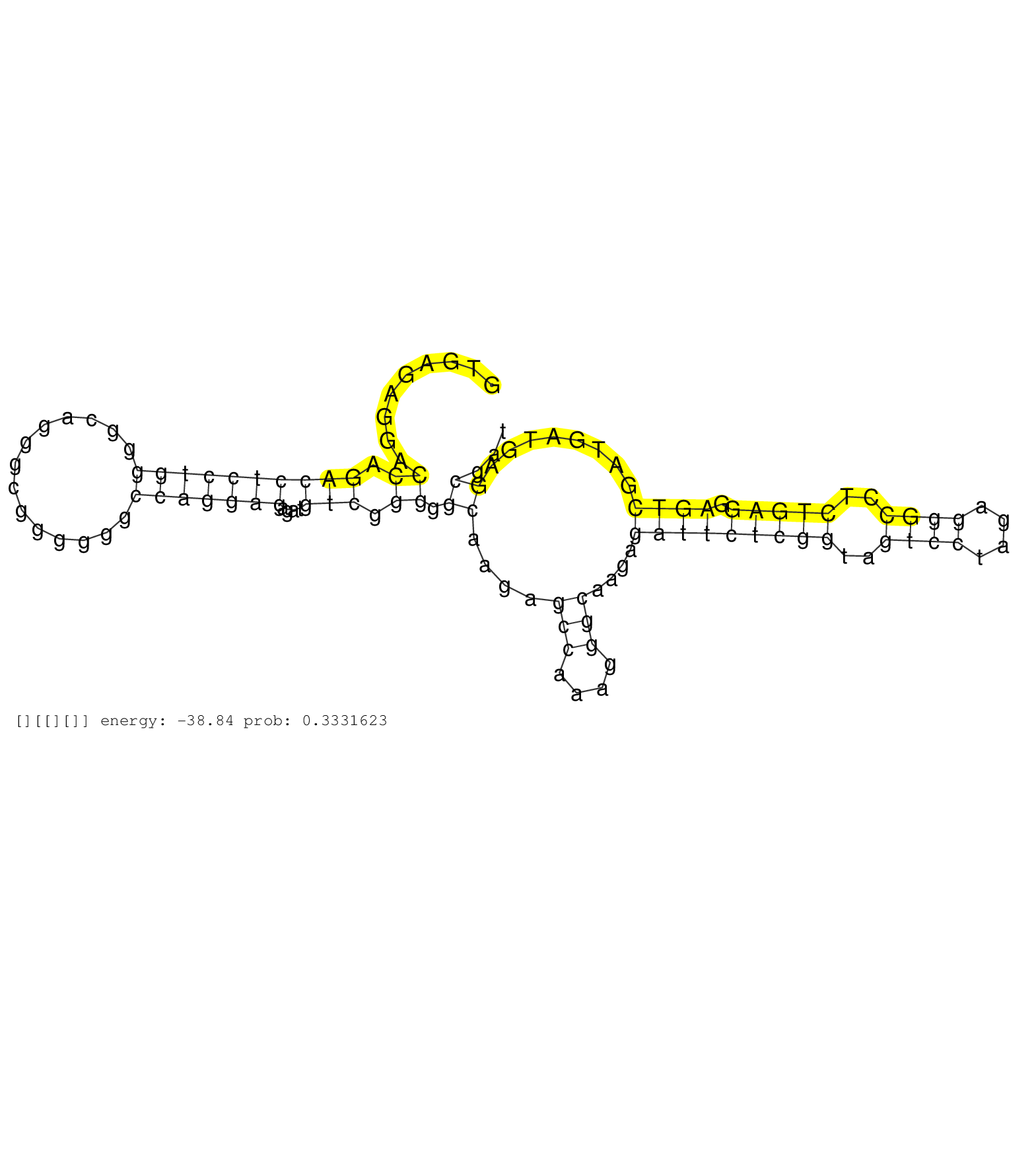
| TATTCTGGTTTTCATAATTTGCGTATATAAGCATTTTATACATGTTATCAGTATCTTCATGTCATTTCAGTTAGTTTTTTAAAAATCAGTTTAACTTTCTAACACTTTGAATCTACATTTTTGTGGGTGTTTGAATTTAACACAATTTAATTTTTCTTTTTTCTGTTGTGTGTATATGTGTCTGTGTGTTACTCTTTCAGTATTTATTAGAGATGAAAAGTTTAATTTTACCTCCAGAACACATTCTGAA ........................................................................................(((.....(((.(((((.......(((((....)))))))))).)))...)))............................................................................................................. ................................................................................81.........................................................................156............................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................TCAGTTTAACTTTCTAACAGA................................................................................................................................................ | 21 | 21.00 | 0.00 | 21.00 | - | - | - | - | - | - | - | - | - | |
| .........................................................................................TTTAACTTTCTAACAGA................................................................................................................................................ | 17 | 18.00 | 0.00 | 18.00 | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................TGAATTTAACACAATTTAATT.................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATTTAATTTTTCTTTTTTCAGT.................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | |
| .................................................................................................................................................................................................CTTTCAGTATTTATTAGAGATGAAAA............................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................AATTTTTCTTTTTTCTGCAG.................................................................................. | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | |
| ........................................................................................................................................................TTTCTTTTTTCTGTTGCTAT.............................................................................. | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...............................................................................................................................................AATTTAATTTTTCTTTTGAG....................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................TATATGTGTCTGTGTGTAG........................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | |
| .........................................................................................TTTAACTTTCTAACAGC................................................................................................................................................ | 17 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | |
| .......................................................................................AGTTTAACTTTCTAACAGA................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................ATTTTTCTTTTTTCTGTTGGTC............................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................CTTTTTTCTGTTGTGTGTATATGTGTCCGTG................................................................ | 31 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................TGTGTATATGTGTCTGCATC.............................................................. | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | |
| ........................................................................AGTTTTTTAAAAATCAGGTAC............................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | |
| ....................................................................................................................................................AATTTTTCTTTTTTCTGTTGACTG.............................................................................. | 24 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | |
| .....................................................................................................................................AATTTAACACAATTTAATTTTTCTTTTTTCCG..................................................................................... | 32 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| TATTCTGGTTTTCATAATTTGCGTATATAAGCATTTTATACATGTTATCAGTATCTTCATGTCATTTCAGTTAGTTTTTTAAAAATCAGTTTAACTTTCTAACACTTTGAATCTACATTTTTGTGGGTGTTTGAATTTAACACAATTTAATTTTTCTTTTTTCTGTTGTGTGTATATGTGTCTGTGTGTTACTCTTTCAGTATTTATTAGAGATGAAAAGTTTAATTTTACCTCCAGAACACATTCTGAA ........................................................................................(((.....(((.(((((.......(((((....)))))))))).)))...)))............................................................................................................. ................................................................................81.........................................................................156............................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189782 | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|