| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (1) AGO2.ip | (1) AGO3.ip | (12) B-CELL | (3) BRAIN | (2) BREAST | (17) CELL-LINE | (1) CERVIX | (2) HEART | (1) HELA | (4) LIVER | (1) OTHER | (1) RRP40.ip | (4) SKIN |
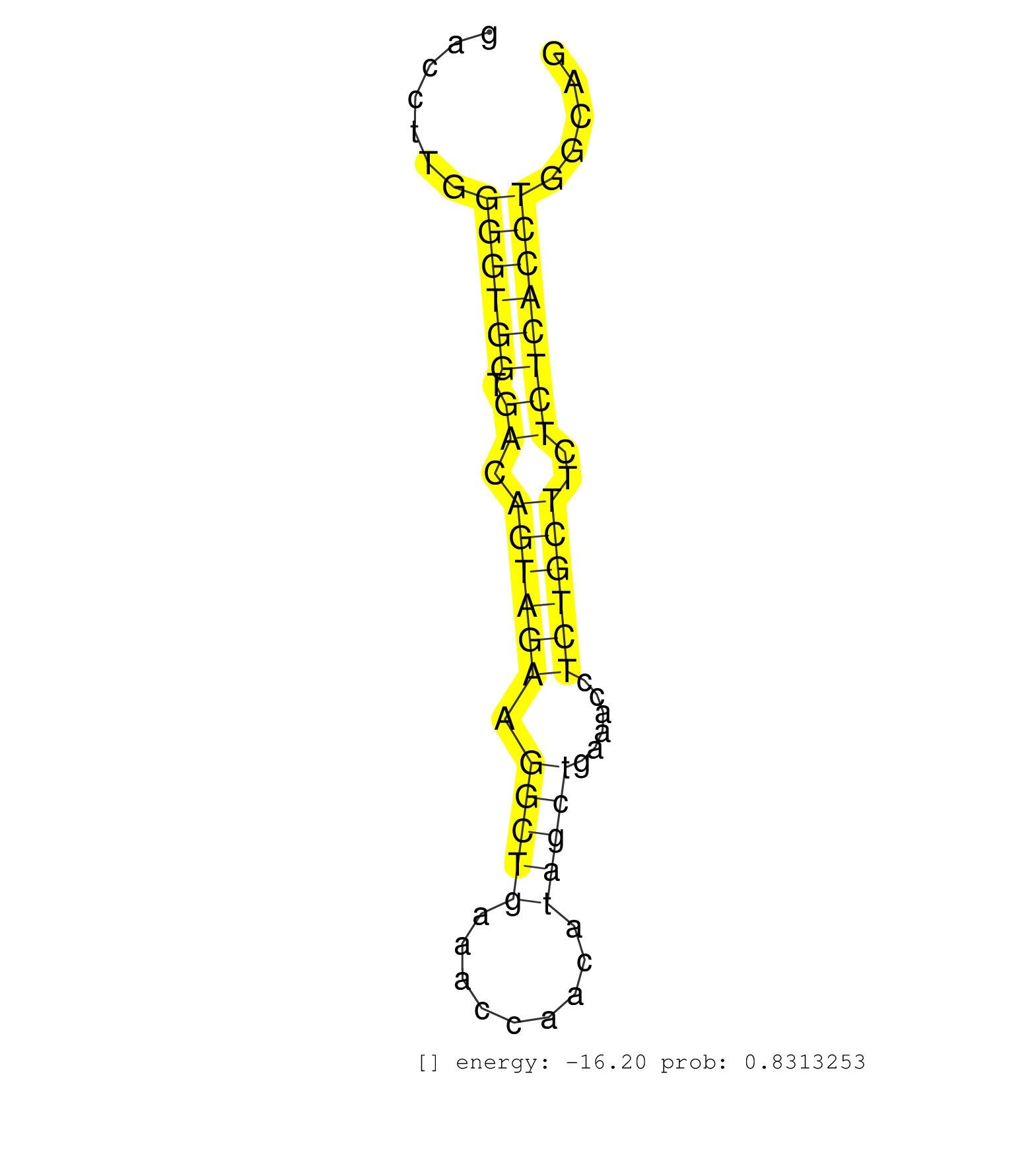
| CTGCCCACCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGTTCATCTTCCATCTTTTCACCTCCACTGCATAGATGTGCCTGTTATATTTCTGAGCCCCCTTCCCCCAGCATTGACCTTGGGGTGGTGACAGTAGAAGGCTGAAACCAACATAGCTGAAACCTCTGCTTCTCTCACCTGGCAGGTAAAAGCAGCTGTTAAGTATGCCCTTAGCGTAGGCTACCGCCACATTGA .........................................................................................................................................((((((.((.((((((.(((((.........)))))......))))))..))))))))....................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR191501(GSM715611) 8genomic small RNA (size selected RNA from to. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR029128(GSM416757) H520. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038860(GSM458543) MM426. (cell line) | SRR038857(GSM458540) D20. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR038863(GSM458546) MM603. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGCT............................................................................................ | 23 | 1 | 34.00 | 34.00 | 16.00 | - | - | - | - | - | 3.00 | 1.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGCT............................................................................................ | 24 | 1 | 11.00 | 11.00 | - | - | - | 1.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGC............................................................................................. | 22 | 1 | 7.00 | 7.00 | 1.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGC............................................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGCTGA.......................................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GGGGTGGTGACAGTAGAAGGC............................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGCTAT.......................................................................................... | 25 | 1 | 2.00 | 34.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGCTT........................................................................................... | 24 | 1 | 2.00 | 34.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGG.............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGCTTT.......................................................................................... | 26 | 1 | 2.00 | 11.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGCTA........................................................................................... | 25 | 1 | 2.00 | 11.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGCTGAAA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAGGCTTG.......................................................................................... | 25 | 1 | 1.00 | 34.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TAGCGTAGGCTACCGCCACAT... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGAAG............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ACAGGCGTGAGCCACCGCGCGGCA................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGA................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................TCTGCTTCTCTCACCTGGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GTGGTGACAGTAGAAGGC............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAAGGCAT........................................................................................... | 25 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGCCCTTAGCGTAGGCTACCGCCACAT... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGGTGGTGACAGTAGACGGA............................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................AAGGCTGAAACCAACATAGCTGAAACCTCTGC.................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TTGGGGTGGTGACAGTAGAA................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GTGGTGACAGTAGAAGGCTGAAACAAAC................................................................................... | 28 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................GCTGTTAAGTATGCCCTTAGCG................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................CTTCTCTCACCTGGCAG.................................................. | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| CTGCCCACCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGTTCATCTTCCATCTTTTCACCTCCACTGCATAGATGTGCCTGTTATATTTCTGAGCCCCCTTCCCCCAGCATTGACCTTGGGGTGGTGACAGTAGAAGGCTGAAACCAACATAGCTGAAACCTCTGCTTCTCTCACCTGGCAGGTAAAAGCAGCTGTTAAGTATGCCCTTAGCGTAGGCTACCGCCACATTGA .........................................................................................................................................((((((.((.((((((.(((((.........)))))......))))))..))))))))....................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207114(GSM721076) "IP against AGO 1 & 2, RRP40 knockdown". (ago1/2 RRP40 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR191501(GSM715611) 8genomic small RNA (size selected RNA from to. (breast) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR029128(GSM416757) H520. (cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038860(GSM458543) MM426. (cell line) | SRR038857(GSM458540) D20. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR038863(GSM458546) MM603. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TGCCCACCTCAGCCTCCCACGAC.................................................................................................................................................................................................................................. | 23 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................AGTTCATCTTCCATCTTATT............................................................................................................................................................................... | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .TGCCCACCTCAGCCTCCCGA..................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................TTACAGGCGTGAGCCACCGCGCCCAGTTCCCC........................................................................................................................................................................................... | 32 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................GGCGTGAGCCACCGCGCCCAGTCG.............................................................................................................................................................................................. | 24 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................CAGGCGTGAGCCACCGCGACA................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .TGCCCACCTCAGCCTCCCCGCA................................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CCTTCTACTGTCACCA.............................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |