| (1) AGO2.ip | (1) B-CELL | (4) BREAST | (14) CELL-LINE | (1) FIBROBLAST | (2) HEART | (1) HELA | (1) LIVER | (13) SKIN | (2) XRN.ip |
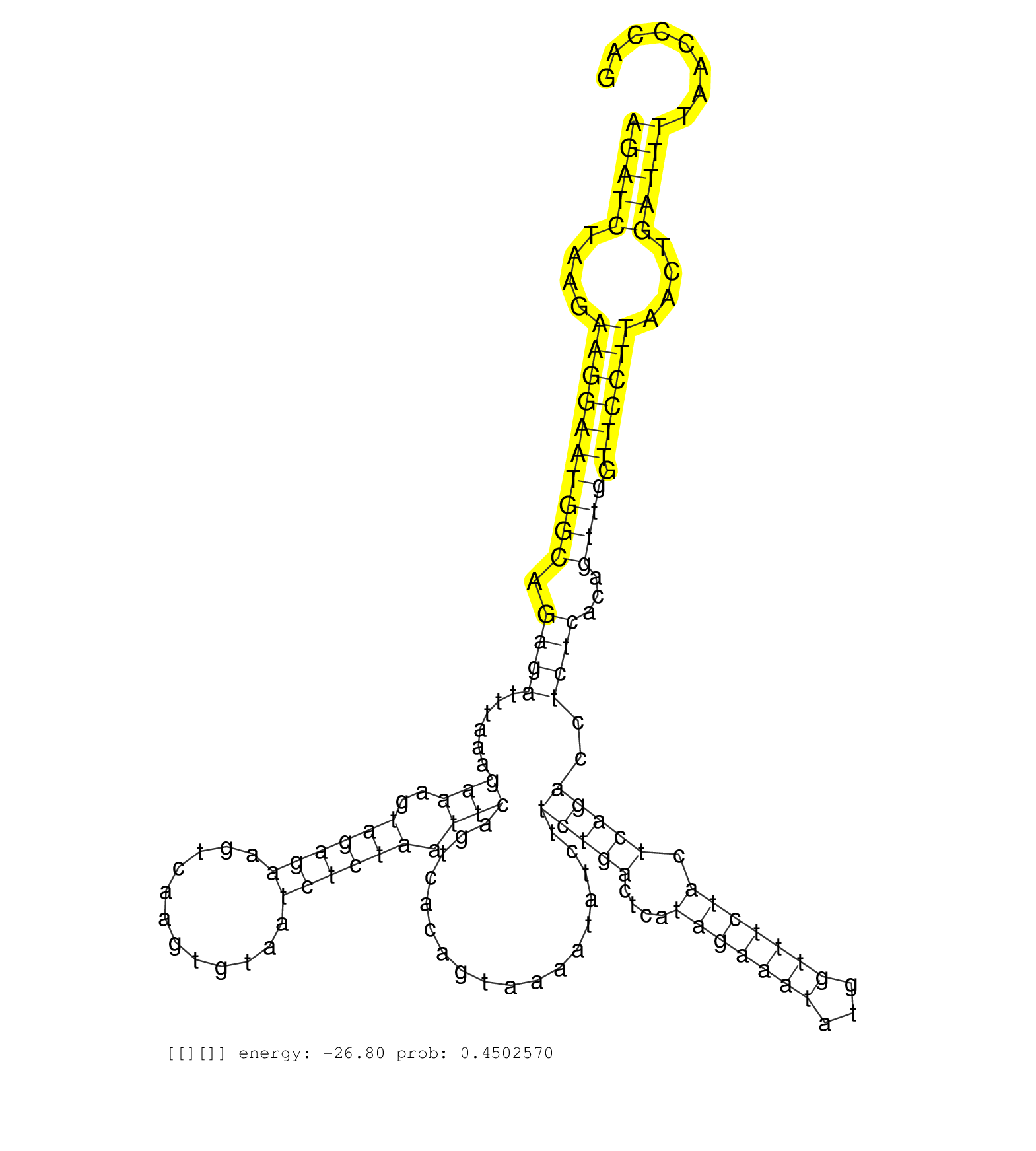
| AAGGTTTTTGAACTTTGAAATAATATAAACTCATTTTTTTTAAGTAGTAGAGATCTAAGAAGGAATGGCAGAGATTTAAAGAAAGTAGAGAAGTCAAGTGTAATCTCTAATTCAGTCACAGTAAAATATCTTCTGACTCATAGAAATATGGTTTCTACTCAGACCTCTCACAGTTGGTTCCTTAACTGATTTTAACCCAGGTACCTGATTTCCCTCTCAGGAGCTCTGGCTATTGTTAATGCAGTACCCT ..................................................(((((....((((((((((.((((......(((..((((((............)))))).)))..................(((((....(((((((...))))))).)))))..))))...)))).))))))....))))).......................................................... ..................................................51...................................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................GTTCCTTAACTGATTTTAACCCAG.................................................. | 24 | 1 | 34.00 | 34.00 | 6.00 | 10.00 | - | - | - | - | - | - | - | - | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 |
| ................................................................................................................................................................................GTTCCTTAACTGATTTTAACCCAGA................................................. | 25 | 1 | 26.00 | 34.00 | 22.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACTGATTTTAACCCAGAA................................................ | 25 | 1 | 14.00 | 5.00 | - | - | - | - | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACTGATTTTAACCCAGA................................................. | 24 | 1 | 8.00 | 5.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACTGATTTTAACCCAG.................................................. | 23 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTTAACTGATTTTAACCCAGAA................................................ | 22 | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................GTAGAGATCTAAGAAGGAATGGCAG................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GGTTCCTTAACTGATTTTAACCCAGG................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GGTTCCTTAACTGATTTTAACCCAGA................................................. | 26 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................CAGGAGCTCTGGCTATTGTT............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................TGGTTCCTTAACTGATTTTAACCCAGA................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................GAATGGCAGAGATTTAAAGAAAGTAGAGAAG............................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................GGAGCTCTGGCTATTGTTAATGCAGTACC.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................TCTAAGAAGGAATGGCAGAGATTTAAAGAAAGTAGA................................................................................................................................................................. | 36 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................GTTCCTTAACTGATTTTAACCCAGAG................................................ | 26 | 1 | 1.00 | 34.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CTCTCAGGAGCTCTGGCTATT................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................AAGAAGGAATGGCAGAAGAG.............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................TCTCAGGAGCTCTGGCTAT................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACTGATTTTAACCCAGG................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GTAGTAGAGATCTAAGAAGGAATGG...................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| AAGGTTTTTGAACTTTGAAATAATATAAACTCATTTTTTTTAAGTAGTAGAGATCTAAGAAGGAATGGCAGAGATTTAAAGAAAGTAGAGAAGTCAAGTGTAATCTCTAATTCAGTCACAGTAAAATATCTTCTGACTCATAGAAATATGGTTTCTACTCAGACCTCTCACAGTTGGTTCCTTAACTGATTTTAACCCAGGTACCTGATTTCCCTCTCAGGAGCTCTGGCTATTGTTAATGCAGTACCCT ..................................................(((((....((((((((((.((((......(((..((((((............)))))).)))..................(((((....(((((((...))))))).)))))..))))...)))).))))))....))))).......................................................... ..................................................51...................................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR314796(SRX084354) "Total RNA, fractionated (15-30nt)". (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................AATATGGTTTCTACTATC........................................................................................ | 18 | 9.00 | 0.00 | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................AGTAGAAACCATATTT........................................................................................... | 16 | 0 | 7.00 | 7.00 | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |