| (5) B-CELL | (3) BRAIN | (20) BREAST | (31) CELL-LINE | (4) CERVIX | (3) FIBROBLAST | (3) HEART | (2) HELA | (1) KIDNEY | (6) LIVER | (2) OTHER | (1) RRP40.ip | (41) SKIN | (1) TESTES | (4) UTERUS | (2) XRN.ip |
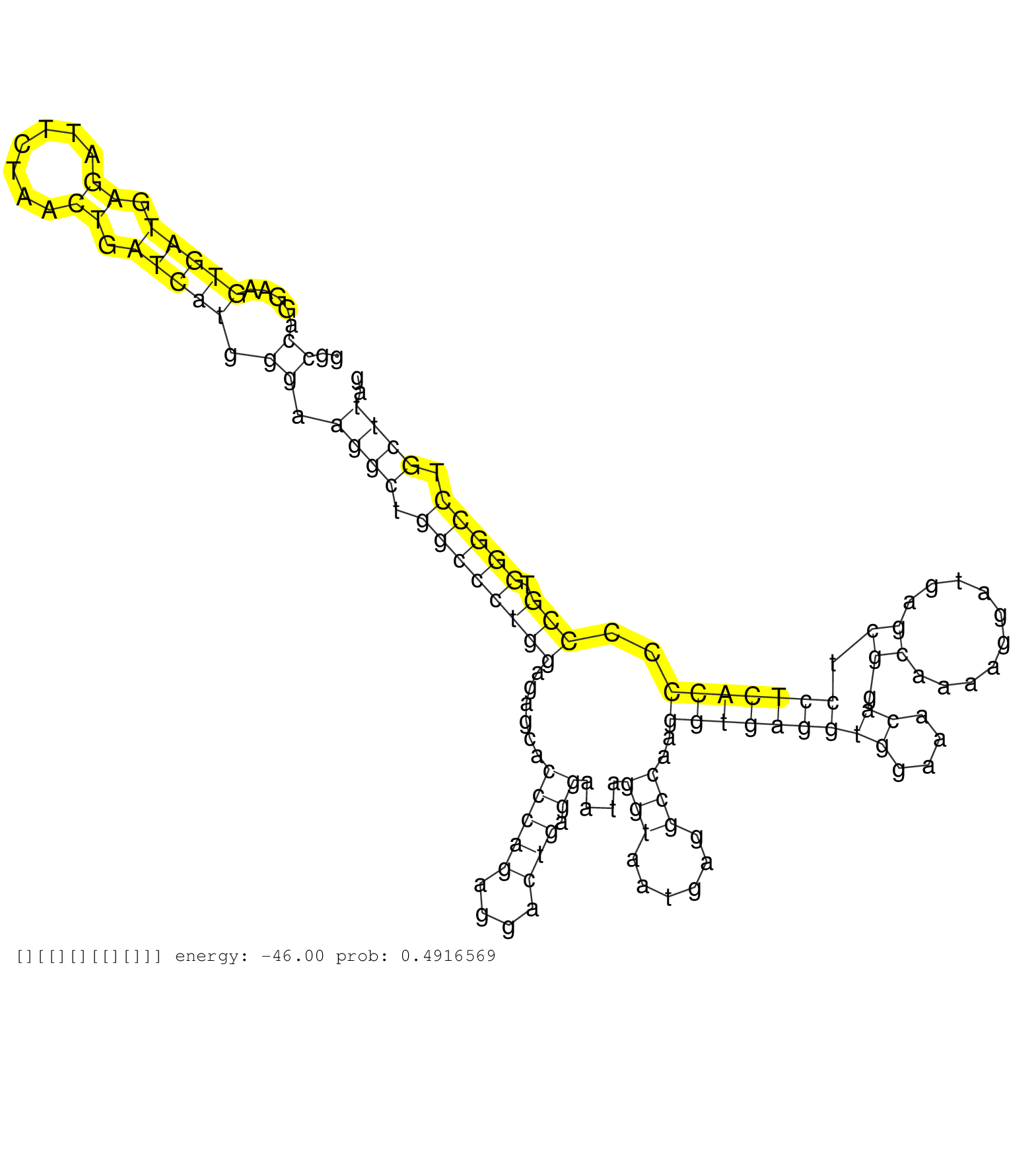
| TCACGGTTTAGGAAATTGAGGCTTTGAGAGGTGAAATCGCTTGCCTTAGAGTCCTGGGCCAGGAAGTGATGAGATTCTAACTGATCATGGGAAGGCTGGCCCTGGAGAGCACCCAGAGGACTGAGGAATAGGTAATGAGGCCAAGGTGAGGTGGAAACAGGCAAAAGGATGAGCTCCTCACCCCCCGTGGGCCTGCTTAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGG ..........................................................((.....(((((.((.......)).))))).)).((((.((((((((......(((((....))).))....(((......)))..(((((((((....)).((..........)).)))))))..))).))))).)))).................................................... ........................................................57.............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM416733(GSM416733) HEK293. (cell line) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR037938(GSM510476) 293Red. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191597(GSM715707) 75genomic small RNA (size selected RNA from t. (breast) | SRR038859(GSM458542) MM386. (cell line) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR189784 | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038854(GSM458537) MM653. (cell line) | SRR191627(GSM715737) 45genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040034(GSM532919) G001N. (cervix) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191480(GSM715590) 38genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................GGAAGTGATGAGATTCTAACTGATC.................................................................................................................................................................... | 25 | 1 | 357.00 | 357.00 | 54.00 | 58.00 | 41.00 | 22.00 | 50.00 | 37.00 | - | 11.00 | 4.00 | 10.00 | 14.00 | 3.00 | 2.00 | 4.00 | - | 2.00 | 4.00 | - | - | 1.00 | 6.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 3.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCA................................................................................................................................................................... | 27 | 1 | 137.00 | 137.00 | 19.00 | 18.00 | - | 20.00 | - | - | 6.00 | - | 1.00 | 1.00 | 4.00 | 2.00 | 3.00 | 1.00 | - | - | - | 2.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | 4.00 | 4.00 | - | 1.00 | 3.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATCA................................................................................................................................................................... | 26 | 1 | 130.00 | 130.00 | 20.00 | 15.00 | 24.00 | 8.00 | 11.00 | 18.00 | - | 24.00 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGTGATGAGATTCTAACTGATC.................................................................................................................................................................... | 22 | 1 | 120.00 | 120.00 | 23.00 | 19.00 | 12.00 | 16.00 | 2.00 | 15.00 | - | 13.00 | 5.00 | - | - | - | 5.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATC.................................................................................................................................................................... | 26 | 1 | 116.00 | 116.00 | - | - | 7.00 | 5.00 | - | 1.00 | 44.00 | - | 4.00 | 2.00 | - | 10.00 | 1.00 | - | 3.00 | - | 2.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 2.00 | - | - | - | 4.00 | - | - | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCAT.................................................................................................................................................................. | 28 | 1 | 61.00 | 61.00 | 9.00 | 8.00 | 13.00 | 10.00 | 5.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATCAT.................................................................................................................................................................. | 27 | 1 | 37.00 | 37.00 | - | - | 8.00 | 1.00 | 17.00 | - | - | - | 3.00 | - | 2.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCATG................................................................................................................................................................. | 29 | 1 | 29.00 | 29.00 | - | - | 6.00 | 13.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATCATG................................................................................................................................................................. | 28 | 1 | 18.00 | 18.00 | 2.00 | 3.00 | - | 6.00 | 1.00 | 3.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GTGATGAGATTCTAACTGATC.................................................................................................................................................................... | 21 | 1 | 15.00 | 15.00 | - | - | - | 5.00 | - | 9.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGA...................................................................................................................................................................... | 24 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGAGATTCTAACTGATCA................................................................................................................................................................... | 18 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTG....................................................................................................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGAT..................................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGAT..................................................................................................................................................................... | 24 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTG....................................................................................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTTATC.................................................................................................................................................................... | 25 | 4.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................GGAAGTGATGAGATTCTAACTGA...................................................................................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GAAGTGATGAGATTCTAACTGATC.................................................................................................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TCACCCCCCGTGGGCCTG....................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GTGATGAGATTCTAACTGATCATA................................................................................................................................................................. | 24 | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................CTGATCATGGGAAGGCTGGCC..................................................................................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATCAA.................................................................................................................................................................. | 27 | 1 | 3.00 | 130.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATA.................................................................................................................................................................... | 25 | 1 | 2.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGCTC.................................................................................................................................................................... | 25 | 1 | 2.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAAC......................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACT........................................................................................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGGTC.................................................................................................................................................................... | 25 | 1 | 2.00 | 5.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ACTGATCATGGGAAGGCTGGCC..................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................TAACTGATCATGGGAAGGCTGGCCCTGG................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................ATGAGATTCTAACTGATCAT.................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGATT.................................................................................................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GATCATGGGAAGGCTGGCC..................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGAGGACTGAGGAATAGGTA.................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GGAGAGCACCCAGAGGACTGAGGAATA........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CCCAGAGGACTGAGGAATAGGT..................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CAGAGGACTGAGGAATAGGTAATG................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGAGGACTGAGGAATAGGT..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................AACTGATCATGGGAAGGCTGGCT..................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................AGTGATGAGATTCTAACTGAT..................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGTGATGAGATTCTAACTGATCAT.................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGACC.................................................................................................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATTA................................................................................................................................................................... | 27 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CAGAGGACTGAGGAATAGGTAATGAGGCC............................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................AACTGATCATGGGAAGGCTGGCCC.................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGGTGAGGTGGAAACAGGCAAAAGGA................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCATGG................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCAAAAA............................................................................................................................................................... | 31 | 1 | 1.00 | 137.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................TGGAAACAGGCAAAAATTG................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCATAAA............................................................................................................................................................... | 31 | 1 | 1.00 | 61.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CAGAGGACTGAGGAATAGGTA.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAACTGTTC.................................................................................................................................................................... | 25 | 1 | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAAC......................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ATGAGGCCAAGGTGAACCG................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................GAGGACTGAGGAATAGGTAATGAGGCCA........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCAAG................................................................................................................................................................. | 29 | 1 | 1.00 | 137.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGATCATA................................................................................................................................................................. | 29 | 1 | 1.00 | 61.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GGTTTAGGAAATTGAGGCTTTGAGAGGTG......................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTCTAACTGCTCA................................................................................................................................................................... | 27 | 1 | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGAAGTGATGAGATTCTAATT........................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................AGTGATGAGATTCTAACTGATCATG................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GAGGACTGAGGAATAGGTAAT.................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGTGATGAGATTCTAACTGATCA................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................CAGGCAAAAGGATGAGCTCCTCACCCC.................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GAGGACTGAGGAATAGGTAATGAGGC............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CGGTTTAGGAAATTGAGGCTTTGAGAGGG.......................................................................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................AGGAAGTGATGAGATTCAC........................................................................................................................................................................... | 19 | 2 | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AGGAAGTGATGAGATTC............................................................................................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ........................................................................................................GAGAGCACCCAGAGGACTGAG............................................................................................................................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCACGGTTTAGGAAATTGAGGCTTTGAGAGGTGAAATCGCTTGCCTTAGAGTCCTGGGCCAGGAAGTGATGAGATTCTAACTGATCATGGGAAGGCTGGCCCTGGAGAGCACCCAGAGGACTGAGGAATAGGTAATGAGGCCAAGGTGAGGTGGAAACAGGCAAAAGGATGAGCTCCTCACCCCCCGTGGGCCTGCTTAGTCTCCCCTCATCAGGACGCCTCCTTCCTGTACACGGAGCCCCTGGGCCGG ..........................................................((.....(((((.((.......)).))))).)).((((.((((((((......(((((....))).))....(((......)))..(((((((((....)).((..........)).)))))))..))).))))).)))).................................................... ........................................................57.............................................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR015447(SRR015447) nuclear small RNAs. (breast) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM416733(GSM416733) HEK293. (cell line) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR037938(GSM510476) 293Red. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR029130(GSM416759) DLD2. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191597(GSM715707) 75genomic small RNA (size selected RNA from t. (breast) | SRR038859(GSM458542) MM386. (cell line) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR189784 | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038854(GSM458537) MM653. (cell line) | SRR191627(GSM715737) 45genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191414(GSM715524) 31genomic small RNA (size selected RNA from t. (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | GSM359208(GSM359208) hepg2_bindASP_hl_2. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR040034(GSM532919) G001N. (cervix) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR029124(GSM416753) HeLa. (hela) | TAX577580(Rovira) total RNA. (breast) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191480(GSM715590) 38genomic small RNA (size selected RNA from t. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM532874(GSM532874) G699T. (cervix) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR191596(GSM715706) 73genomic small RNA (size selected RNA from t. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................CGCCTCCTTCCTGTAACTG............... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................GCCTTAGAGTCCTGGCC............................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ......................................GCTTGCCTTAGAGTCCAGC................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................GACGCCTCCTTCCTGAGAA................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................AAGGATGAGCTCCTCACCGAT................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................ATTTCACCTCTCAAAG..................................................................................................................................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |