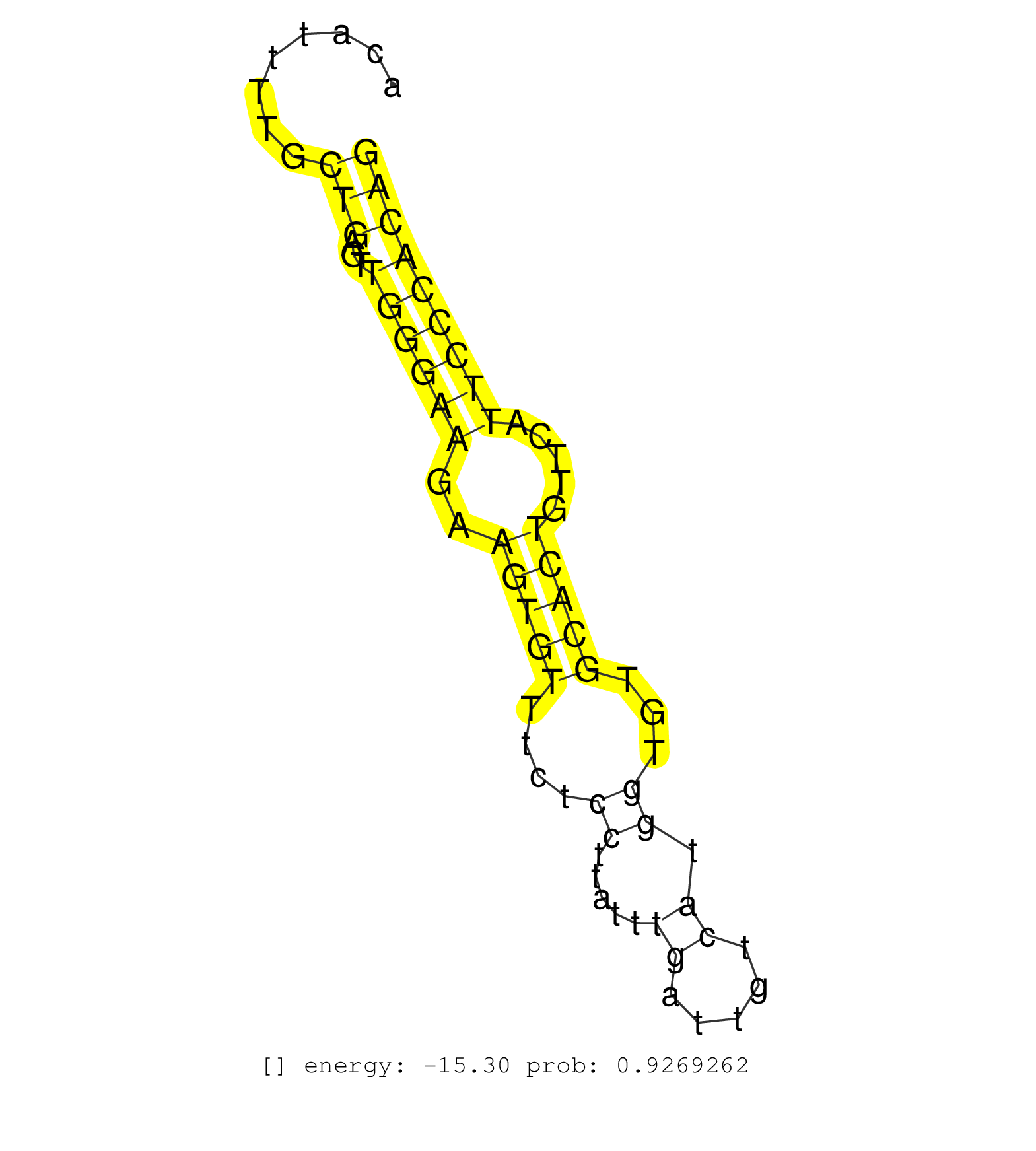
| (1) B-CELL | (3) BREAST | (8) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (3) HELA | (2) LIVER | (1) OTHER | (27) SKIN | (1) XRN.ip |
| ATGAGTGCTTCAGTCCTTCATATTGCCAAATACTCTATTGCATGTGTATGCTGCATTTTGTTTATCCATTCACAGTTGATGGACACTTGAAGTGTTCTTGCTTTCTGGCAGTTAGGAATAATGCTGCTGTGGACCTACATTTTTGTGTGAACATATGTTTTCATTTCTCATTTTTGTTTCCTCTTGTGCATTAACTGTAGGAACTCAATGTTCGAAAAGTTACACTGTCTACAGATAAAAACAAGTATGG .........................................................................(((....((((((.....))))))..)))....((((((..........)))))).......................................................................................................................... ...................................................................68...........................................................129....................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR444061(SRX128909) Sample 19cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR191614(GSM715724) 92genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR189785 | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM359178(GSM359178) hela_nucl_t. (hela) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................GCAGTTAGGAATAATGGA............................................................................................................................. | 18 | 1 | 13.00 | 1.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................GCAGTTAGGAATAATGG.............................................................................................................................. | 17 | 1 | 7.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ............................................................................................................CAGTTAGGAATAATGGAA............................................................................................................................ | 18 | 7.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................CAGTTAGGAATAATGGAAT........................................................................................................................... | 19 | 7.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................GCAGTTAGGAATAATGGAAT........................................................................................................................... | 20 | 1 | 5.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................GCAGTTAGGAATAATGGAA............................................................................................................................ | 19 | 1 | 5.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................ACAGATAAAAACAAGTATGG | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AAGTTACACTGTCTACAGA............... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................GCAGTTAGGAATAATG............................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTGCTTCAGTCCTTCCGTC................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CTCAATGTTCGAAAAGTTACACTGTCTACAGATA............. | 34 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CAGTTAGGAATAATGGAT............................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................ATGCTGCTGTGGACCTACATTTTG.......................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................AAGTTACACTGTCTACAGATA............. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CAGTTAGGAATAATGTA............................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................CAGTTGATGGACACTATG................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................TTTTCATTTCTCATTTTTGTTTCCTCTCTTA.............................................................. | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .................................................................................................................................................................................................................GTTCGAAAAGTTACACTGTCTACAGATAAAAAC........ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTGCTTCAGTCCTTCGA..................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................CATTTTTGTTTCCTCTTGTGCAT........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GTGCTTCAGTCCTTCTATC................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................ATTGCATGTGTATGCTGCATTTTGTT............................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....GTGCTTCAGTCCTTCTA..................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCCTCTTGTGCATTAACTGTAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AAGTTACACTGTCTACAGAT.............. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TTTTCATTTCTCATTTTTTTG........................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ................................................................................................................................................................................................................TGTTCGAAAAGTTACACT........................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CTCAATGTTCGAAAAGTTACACTGTCTAC.................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTTTGTTTCCTCTTGTGC............................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ATGAGTGCTTCAGTCCTTCATATTGCCAAATACTCTATTGCATGTGTATGCTGCATTTTGTTTATCCATTCACAGTTGATGGACACTTGAAGTGTTCTTGCTTTCTGGCAGTTAGGAATAATGCTGCTGTGGACCTACATTTTTGTGTGAACATATGTTTTCATTTCTCATTTTTGTTTCCTCTTGTGCATTAACTGTAGGAACTCAATGTTCGAAAAGTTACACTGTCTACAGATAAAAACAAGTATGG .........................................................................(((....((((((.....))))))..)))....((((((..........)))))).......................................................................................................................... ...................................................................68...........................................................129....................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR444061(SRX128909) Sample 19cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR191614(GSM715724) 92genomic small RNA (size selected RNA from t. (breast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR189782 | SRR189785 | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577741(Rovira) total RNA. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM359178(GSM359178) hela_nucl_t. (hela) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TGAGTGCTTCAGTCCTTCATATTGCCAG............................................................................................................................................................................................................................. | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................CCATCAACTGTGAATG........................................................................................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...............................................GATAAACAAAATGCAGCAT........................................................................................................................................................................................ | 19 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |