| (1) AGO1.ip | (1) BRAIN | (9) CELL-LINE | (1) CERVIX | (5) HEART | (1) LIVER | (4) OTHER | (6) SKIN |
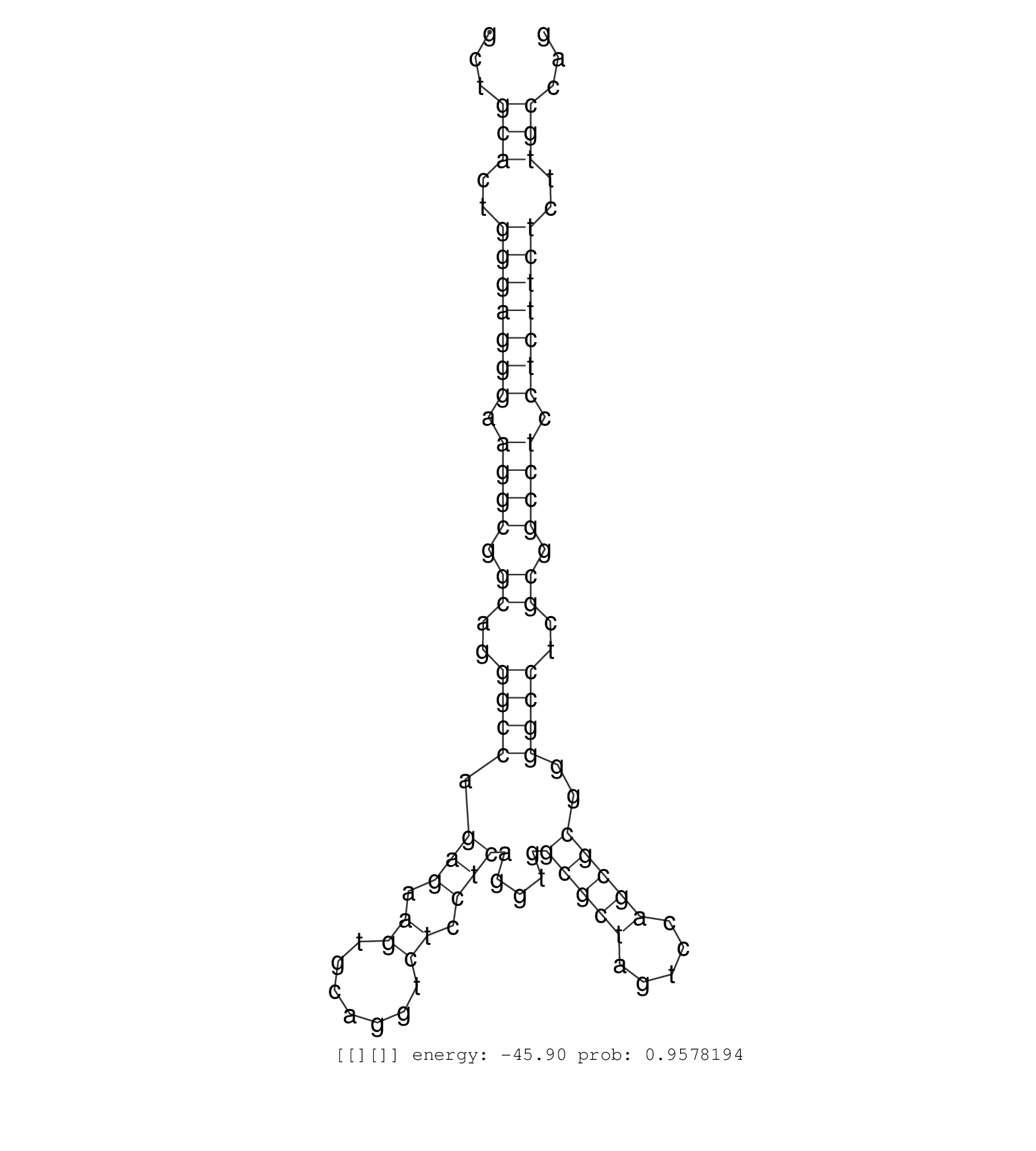
| GGCATGGGCGGGTACACGGGCATGGGGTCTGAGGTCAGGTAGGGTTGCCTCCCTCTGCAGGCCTTGTTTTCCTCCCCCGTGAGGTGGAGGCCACACTGCTCACCTGCTAGGAGCCACAGGGAGGAGCCACTGTGGGAGCGGGGAAGAGGCTCATGCAGGGCCTGGCCCAGGAGAGTGACACGTCCTGTCTCCGTCTTTAGGTGGGAGAAGGAGAAGCACGAAGACGGGGTGAAGTGGAGACAGCTGGAGC ............................................................................((....))....((((......((..((((((.....((((((((.....)).)))))).))))))..)).(((((.....))))).))))..(((((..((....))...))))).......................................................... ............................................................................77.........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR189785 | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR040036(GSM532921) G243N. (cervix) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189782 | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................AGGTGGAGGCCACACGT........................................................................................................................................................ | 17 | 115.00 | 0.00 | 41.00 | 30.00 | 24.00 | 8.00 | 6.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ............................................................................CCGTGAGGTGGAGGCGG............................................................................................................................................................. | 17 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | |
| ............................................................................CCGTGAGGTGGAGGCG.............................................................................................................................................................. | 16 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | |
| ...................................................................................................................................................................................................................................GGTGAAGTGGAGACAGCT..... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................AGGTGGAGGCCACACGC........................................................................................................................................................ | 17 | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ............................CTGAGGTCAGGTAGGG.............................................................................................................................................................................................................. | 16 | 3 | 1.33 | 1.33 | - | - | - | - | - | - | - | - | - | 1.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGACACGTCCTGTCTCCGTCTTTA................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GAGGAGCCACTGTGGGATT............................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................ACGGGGTGAAGTGGAGAC......... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGGGAGGAGCCACTGTAGCC................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................CTGAGGTCAGGTAGGGTTG........................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................................AGACGGGGTGAAGTGGAGACAGCTGGAGC | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGGGAGGAGCCACTGTAGCT................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................CGGGGTGAAGTGGAGACAGCCGG... | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................GTCCTGTCTCCGTCTTTAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................AGGTGGAGGCCACACAT........................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................AGGTGGAGGCCACACGA........................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................................GAAGTGGAGACAGCTG.... | 16 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | 0.17 |
| GGCATGGGCGGGTACACGGGCATGGGGTCTGAGGTCAGGTAGGGTTGCCTCCCTCTGCAGGCCTTGTTTTCCTCCCCCGTGAGGTGGAGGCCACACTGCTCACCTGCTAGGAGCCACAGGGAGGAGCCACTGTGGGAGCGGGGAAGAGGCTCATGCAGGGCCTGGCCCAGGAGAGTGACACGTCCTGTCTCCGTCTTTAGGTGGGAGAAGGAGAAGCACGAAGACGGGGTGAAGTGGAGACAGCTGGAGC ............................................................................((....))....((((......((..((((((.....((((((((.....)).)))))).))))))..)).(((((.....))))).))))..(((((..((....))...))))).......................................................... ............................................................................77.........................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189783 | SRR189785 | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR040036(GSM532921) G243N. (cervix) | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189782 | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................CTAGGAGCCACAGGGAACAA............................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |