| (1) AGO2.ip | (3) B-CELL | (2) BRAIN | (6) BREAST | (9) CELL-LINE | (2) HEART | (1) HELA | (8) LIVER | (1) OTHER | (12) SKIN | (1) TESTES | (4) UTERUS | (1) XRN.ip |
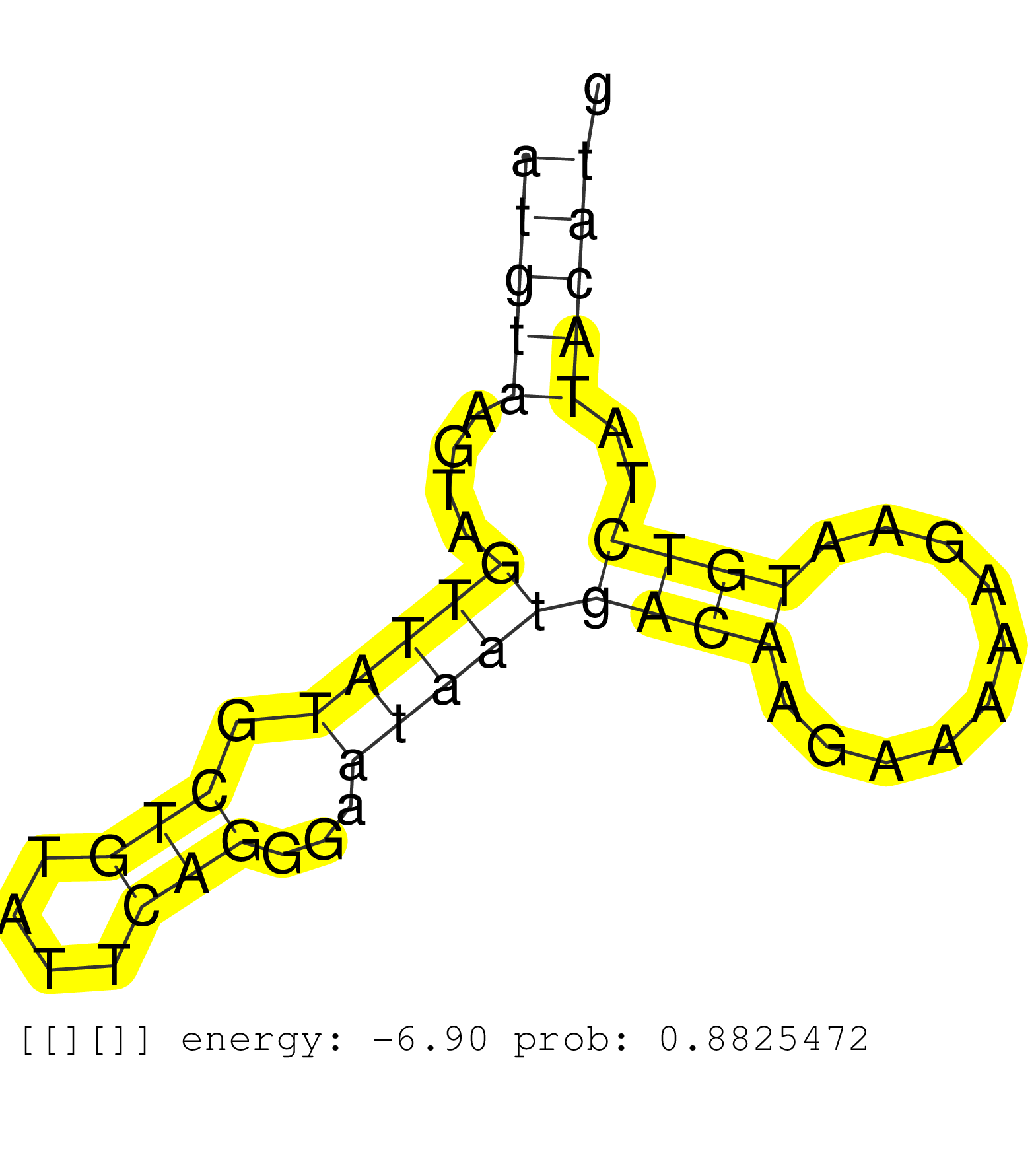
| CTGATATAAAATGGCATAGTATTTGCATATAATCTATGGCATCCTGTATAATTTGTCATCTCTAGATTACTTGTAATACCTAATACAATGTAAATGGTATGTAAGTAGTTATGCTGTATTCAGGGAATAATGACAAGAAAAAGAATGTCTATACATGTGCAGTAGAGACAATTTTTTTTCTGAATACTTTTGATCTACAGTTGATCGAATCTACAGATGCAGAACCCATGGATACAGAGGGCCAGCAGTA .......................................................................................................................................(((((((((((((((.(((.......))).))))).))))))))))..................................................................... ...............................................................................................................................128..........................................................189........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR029130(GSM416759) DLD2. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................TAGAGACAATTTTTTTTCTGAAT................................................................. | 23 | 1 | 10.00 | 10.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACAAGAAAAAGAATGTCTATA................................................................................................. | 21 | 1 | 8.00 | 8.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTGAAT................................................................. | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTGA................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CGAATCTACAGATGCAGAACCCATT.................... | 25 | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................TCTATGGCATCCTGTATAATTTGT.................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTG.................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACAAGAAAAAGAATGTCTATAC................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTCAGGGAATAATGACAAGAAAAAGAATGT...................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCT..................................................................... | 18 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTC...................................................................... | 17 | 7 | 1.29 | 1.29 | - | - | - | 0.14 | - | - | - | - | - | - | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.57 | - | - | - | 0.14 |
| .......................................................................................................AGTAGTTATGCTGTATTCAGGG............................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GCAGAACCCATGGATACAGAGGGCCAGC.... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................ATTTTTTTTCTGAATACTTTTGAT........................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTGAA.................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................ATCGAATCTACAGATGCA............................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTGAAA................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGTAAGTAGTTATGCTGTATTCAGGG............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CTATGGCATCCTGTATAATTTGT.................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CAGGGAATAATGACAAGAAAAAGAATGT...................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CATGTGCAGTAGAGACAATT............................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ATGGCATCCTGTATAATTTG................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACAAGAAAAAGAATGTCTAT.................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GACAATTTTTTTTCTCCTT................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .................................................................................................................................................................GTAGAGACAATTTTTTTTCTGA................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACATGTGCAGTAGAGACA................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CTATACATGTGCAGTAGAGACAATTTTT.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGATCGAATCTACAGATGCAGAACCCA...................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................AGTAGTTATGCTGTATTCAGGGATT.......................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................TATAATTTGTCATCTCTAGATTACTT.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TTTTTCTGAATACTTTTGATCTACAG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTATACATGTGCAGTAGAGACA................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACAAGAAAAAGAATGTCTATACA............................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................AGTTATGCTGTATTCAGG.............................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGAGACAATTTTTTTTCTGAATA................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTATTCAGGGAATAATGACAAGAAAAAG........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................CTACAGATGCAGAACCCATGGATACAG............. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GGGAATAATGACAAGAAAAGGCC......................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AGTTATGCTGTATTCAGGGA............................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................ATGCAGAACCCATGGATACAGAGGGCCA...... | 28 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCAGAACCCATGGATACAGAGGGCC....... | 26 | 4 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................AATACAATGTAAATGGTATGTAAGTAG.............................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................................................................................................................................................................................CAGAACCCATGGATACAGAGGGCCA...... | 25 | 7 | 0.14 | 0.14 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................AGATGCAGAACCCATGGATACAGAGG.......... | 26 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - |
| CTGATATAAAATGGCATAGTATTTGCATATAATCTATGGCATCCTGTATAATTTGTCATCTCTAGATTACTTGTAATACCTAATACAATGTAAATGGTATGTAAGTAGTTATGCTGTATTCAGGGAATAATGACAAGAAAAAGAATGTCTATACATGTGCAGTAGAGACAATTTTTTTTCTGAATACTTTTGATCTACAGTTGATCGAATCTACAGATGCAGAACCCATGGATACAGAGGGCCAGCAGTA .......................................................................................................................................(((((((((((((((.(((.......))).))))).))))))))))..................................................................... ...............................................................................................................................128..........................................................189........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR029130(GSM416759) DLD2. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................CTACAGATGCAGAACCCATGGATACACTTC.......... | 30 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................TGGTATGTAAGTAGTAG........................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................TGTATAGACATTCTTT............................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |