| (1) AGO2.ip | (1) B-CELL | (2) BRAIN | (12) BREAST | (6) CELL-LINE | (5) CERVIX | (1) KIDNEY | (1) OTHER | (4) SKIN | (1) TESTES |
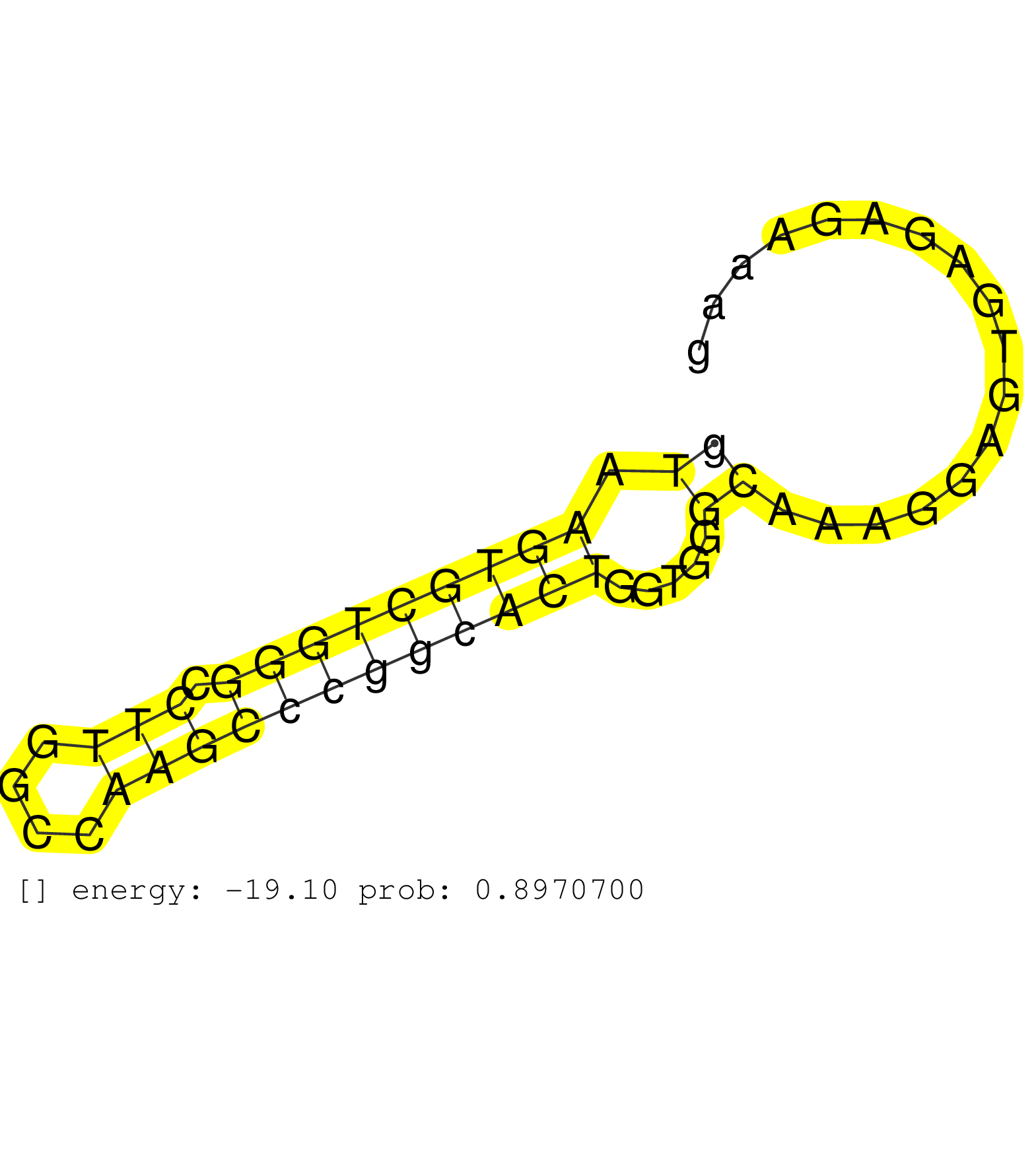
| TCTTGATCTCACTCATCTTCATCTATGAGACCTTCTACAAGCTGGTGAAGGTGGGCAGGCCCCTGCCGAGAGCTGGGCCAGGAGGGCCGAGGTCTCAGGGCTGGAGGGAGTCTCTTGGTCCCATCCTCTGGATTTGAGGCCAGGCTGGTCTGGAGCATCCCCGGGGCTTGTTGCCAACACGTATACCAGGGCTGCAGATTACTCTTTTAATTAATTAATTAATTTGAGACATTCTTACTCTGTTGCCCAG ........................................................................((((((((((..((((..(((((((((.((((((((.......((....)))))))))).))))))))).))))..)))))......)))))...................................................................................... .......................................................................72..........................................................................................164.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | TAX577738(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR040036(GSM532921) G243N. (cervix) | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR189784 | SRR040010(GSM532895) G529N. (cervix) | TAX577746(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577580(Rovira) total RNA. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR444056(SRX128904) Sample 28cDNABarcode: AF-PP-333: ACG CTC TTC . (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR038858(GSM458541) MEL202. (cell line) | SRR040007(GSM532892) G601T. (cervix) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................GCCAGGAGGGCCGAGCGGT........................................................................................................................................................... | 19 | 11.00 | 0.00 | 2.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGCGG............................................................................................................................................................ | 18 | 10.00 | 0.00 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | |
| ............................................................................GCCAGGAGGGCCGAGTGG............................................................................................................................................................ | 18 | 5.00 | 0.00 | - | 1.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTGGT........................................................................................................................................................... | 19 | 4.00 | 0.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGCGGG........................................................................................................................................................... | 19 | 4.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGCG............................................................................................................................................................. | 17 | 4.00 | 0.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGCGC............................................................................................................................................................ | 18 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGCGGA........................................................................................................................................................... | 19 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................TCTCAGGGCTGGAGGGTCCC.......................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................GGCTGGTCTGGAGCACCA.......................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................ACCAGGGCTGCAGATGGT................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGAGG............................................................................................................................................................ | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................GCCGAGGTCTCAGGGCTGG.................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................AGGGCTGCAGATTACTCT............................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GGAGGGAGTCTCTTGTGTC................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................CAGGGCTGGAGGGAGTCGGA....................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....ATCTCACTCATCTTCATC................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................GGAGGGAGTCTCTTGTA................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGAA............................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CACTCATCTTCATCTATG............................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GCCAGGAGGGCCGAGTCGC........................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTGGG........................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTG............................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTAA............................................................................................................................................................ | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTGT............................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................GCCAGGAGGGCCGAGTGGA........................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................TTCTACAAGCTGGTGAAG........................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...........................................................................................................................................................................................AGGGCTGCAGATTAC................................................ | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTTGATCTCACTCATCTTCATCTATGAGACCTTCTACAAGCTGGTGAAGGTGGGCAGGCCCCTGCCGAGAGCTGGGCCAGGAGGGCCGAGGTCTCAGGGCTGGAGGGAGTCTCTTGGTCCCATCCTCTGGATTTGAGGCCAGGCTGGTCTGGAGCATCCCCGGGGCTTGTTGCCAACACGTATACCAGGGCTGCAGATTACTCTTTTAATTAATTAATTAATTTGAGACATTCTTACTCTGTTGCCCAG ........................................................................((((((((((..((((..(((((((((.((((((((.......((....)))))))))).))))))))).))))..)))))......)))))...................................................................................... .......................................................................72..........................................................................................164.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | TAX577738(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR040036(GSM532921) G243N. (cervix) | TAX577740(Rovira) total RNA. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR189784 | SRR040010(GSM532895) G529N. (cervix) | TAX577746(Rovira) total RNA. (breast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577580(Rovira) total RNA. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR444056(SRX128904) Sample 28cDNABarcode: AF-PP-333: ACG CTC TTC . (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR040008(GSM532893) G727N. (cervix) | SRR038858(GSM458541) MEL202. (cell line) | SRR040007(GSM532892) G601T. (cervix) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040025(GSM532910) G613T. (cervix) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR343335 | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR444054(SRX128902) Sample 14cDNABarcode: AF-PP-342: ACG CTC TTC . (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................CTGGAGCATCCCCGGGGGGG................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ........................................................................CTGGGCCAGGAGGGCGTCT............................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................AGGGCCGAGGTCTCAAAAT..................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................CAGGGCTGCAGATTACACT............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |