| (1) AGO1.ip | (1) AGO2.ip | (3) B-CELL | (4) BRAIN | (1) BREAST | (18) CELL-LINE | (1) FIBROBLAST | (3) HEART | (1) KIDNEY | (4) LIVER | (2) OTHER | (14) SKIN |
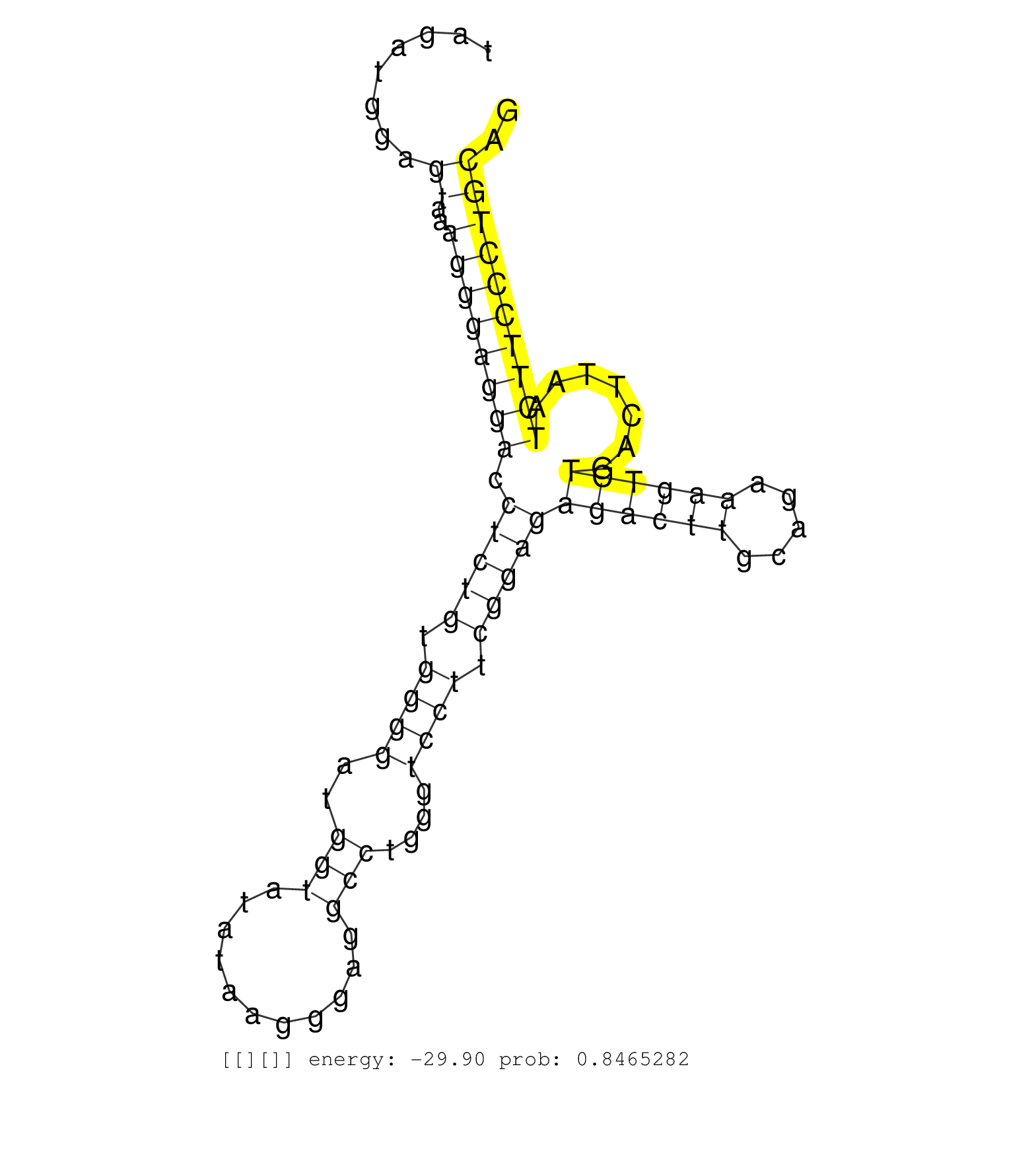
| AGCAGCAAATTATGACCATTATAGATGAACTGGGCAAGGCTTCTGCCAAGGTACTGGAGAGTTTTTAGATGGAGTAAAGGGAGGACCTCTGTGGGGATGGTATATAAGGGAGGCCTGGGTCCTTCGGAGAGACTTGCAGAAAGTCTGACTTAATCTTCCCTGCAGGCCCAGAATCTTTCCGCTCCTATCACTAGTGCATCAAGGATGCAGAGTAA .....................................................((((((...)))))).....((..((((((((.(((((.((((..(((...........)))....)))).)))))((((((.....)))))).......)))))))))).................................................... ..................................................51................................................................................................................165................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189782 | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM339996(GSM339996) hues6Neuron. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR343335 | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR038856(GSM458539) D11. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAG.................................................. | 22 | 1 | 75.00 | 75.00 | 10.00 | 14.00 | 2.00 | 4.00 | 10.00 | 5.00 | 2.00 | - | - | 2.00 | 2.00 | 1.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTG..................................................... | 19 | 1 | 11.00 | 11.00 | - | - | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAGA................................................. | 23 | 1 | 7.00 | 75.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGATGAACTGGGCAAGGCTT............................................................................................................................................................................. | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TCCTTCGGAGAGACTTG............................................................................... | 17 | 2 | 3.50 | 3.50 | - | - | - | - | - | - | - | 3.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TTATAGATGAACTGGGCAAGGCTTCTGCCAAGGC................................................................................................................................................................... | 34 | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGC.................................................... | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAGT................................................. | 23 | 1 | 2.00 | 75.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAT.................................................. | 22 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .......................GATGAACTGGGCAAGGCTTCT........................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TGCCAAGGTACTGGAAG........................................................................................................................................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAA.................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................GAGTTTTTAGATGGAGAC........................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................AACTGGGCAAGGCTTACGT......................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ....................ATAGATGAACTGGGCAAG................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAGTA................................................ | 24 | 1 | 1.00 | 75.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTTCGGAGAGACTTGCAG............................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTGCAAAG................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .................ATTATAGATGAACTGGGCAAGGCTTCTTC......................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................ACTTGCAGAAAGTCTTTT.................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................GAGTAAAGGGAGGACCTCTGTGG......................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCTGACTTAATCTTCCCTTCAG.................................................. | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................CTTCGGAGAGACTTGCAGA........................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ATTATAGATGAACTGGG..................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGATGAACTGGGCAAGGCTTCT........................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................AGGGAGGACCTCTGTGGG........................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GATGAACTGGGCAAG................................................................................................................................................................................. | 15 | 7 | 0.14 | 0.14 | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCAGCAAATTATGACCATTATAGATGAACTGGGCAAGGCTTCTGCCAAGGTACTGGAGAGTTTTTAGATGGAGTAAAGGGAGGACCTCTGTGGGGATGGTATATAAGGGAGGCCTGGGTCCTTCGGAGAGACTTGCAGAAAGTCTGACTTAATCTTCCCTGCAGGCCCAGAATCTTTCCGCTCCTATCACTAGTGCATCAAGGATGCAGAGTAA .....................................................((((((...)))))).....((..((((((((.(((((.((((..(((...........)))....)))).)))))((((((.....)))))).......)))))))))).................................................... ..................................................51................................................................................................................165................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189782 | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM339996(GSM339996) hues6Neuron. (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR343335 | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR029125(GSM416754) U2OS. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189787 | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR038856(GSM458539) D11. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................GGAGAGACTTGCAGAAAG........................................................................ | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................CCTGGGTCCTTCGGAGGA.................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................AAGATTCTGGGCCTGCAG...................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................ATCACTAGTGCATCAGGTC.......... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................AGGCCCAGAATCTTTAGG.................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................GCCTGGGTCCTTCGGATT..................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |