| (3) AGO2.ip | (3) B-CELL | (3) BREAST | (18) CELL-LINE | (4) CERVIX | (4) HEART | (3) LIVER | (2) OTHER | (16) SKIN | (1) TESTES | (1) UTERUS |
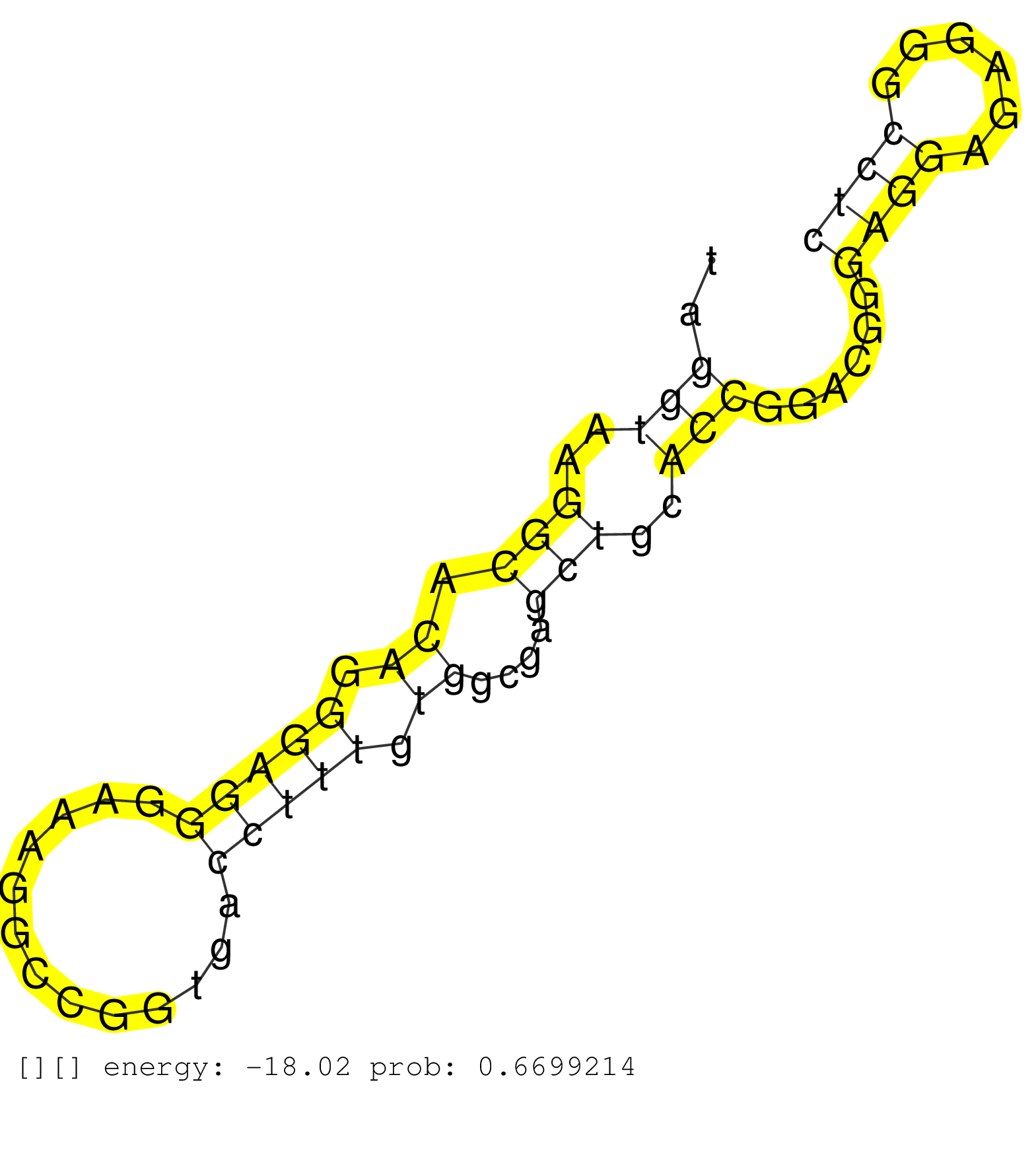
| GAGATCTGGAAGATCAAGAAGCTCATTAAGAGCTTGGAGGCGGCCCGCGGGTGAGCGCCGCGTCCGGCACCCTCCGTGCCCTCTTGGTTCCCGGACCCCTCAGGATGGACGCCCCCGCGCCGCATGGCCGGATGAGGAGAACCAGGCACTAGCCAACTCCCGCCTTCTGGATCTCTGGGTAGTAGCCCCTGACCCAGGGGCGAGGGTCGGCGCGGTTGGCCCGAGAAGGGGTCCTTTCCCCGCATGGGGG ..................................................(((.((((.(((((((((((.....))))).((((((((....))))....)))).))))))....)))))))..(((.(((...((....)).(((....)))...))).))).(((((....)))))....................................................................... ..................................................51...................................................................................................................................184................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040042(GSM532927) G428N. (cervix) | SRR553576(SRX182782) source: Testis. (testes) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR444042(SRX128890) Sample 3cDNABarcode: AF-PP-335: ACG CTC TTC C. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR040043(GSM532928) G428T. (cervix) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR040017(GSM532902) G645T. (cervix) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM956925Ago2(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR444046(SRX128894) Sample 7cDNABarcode: AF-PP-342: ACG CTC TTC C. (skin) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR444051(SRX128899) Sample 11cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................GGGTGAGCGCCGCGTGG......................................................................................................................................................................................... | 17 | 1 | 39.00 | 1.00 | 29.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCGTGG......................................................................................................................................................................................... | 18 | 1 | 19.00 | 2.00 | 7.00 | 4.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCG............................................................................................................................................................................................ | 15 | 2 | 8.50 | 8.50 | 2.50 | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | 0.50 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCGTGGG........................................................................................................................................................................................ | 19 | 1 | 7.00 | 2.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGTGGG........................................................................................................................................................................................ | 18 | 1 | 5.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCGTG.......................................................................................................................................................................................... | 17 | 1 | 4.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CATTAAGAGCTTGGAGGCGGCCCGCGG........................................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TAAGAGCTTGGAGGCAAAA............................................................................................................................................................................................................. | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................CGGGTGAGCGCCGCGT........................................................................................................................................................................................... | 16 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGTGTG........................................................................................................................................................................................ | 18 | 1 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AAGAGCTTGGAGGCGGCCCGCGGCA...................................................................................................................................................................................................... | 25 | 1 | 2.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CCTTCTGGATCTCTGGCA...................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................GCCGCGTCCGGCACCGGG................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................GGGTGAGCGCCGCGTAG......................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CCTTCTGGATCTCTGGGAA..................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................AGAGCTTGGAGGCGGCCCGCGGCA...................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................AAGAGCTTGGAGGCGGCCCGCGGGCG..................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................CGGGTGAGCGCCGCGTGCG........................................................................................................................................................................................ | 19 | 1 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCTGGAAGATCAAGAAGCTCATT............................................................................................................................................................................................................................... | 23 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...ATCTGGAAGATCAAGAAGCT................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CGCGCCGCATGGCCGGATGAT.................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..GATCTGGAAGATCAAGAAGC.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGT........................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCTGGAAGATCAAGAAGC.................................................................................................................................................................................................................................... | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .........................................................................................................................GCATGGCCGGATGAGGAGAACCAGGC....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GCCCCTGACCCAGGGGCGAGGGTCG......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AAGAGCTTGGAGGCGGCCCGCGG........................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGGAAGATCAAGAAGCTCAT................................................................................................................................................................................................................................ | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGTG.......................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CGCGCCGCATGGCCGGATGAG.................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGCCGGATGAGGAGAACCAGGCAC..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGTGGT........................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCGTGGGG....................................................................................................................................................................................... | 20 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................ATGAGGAGAACCAGGCACTAGCCAA.............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................GGGTGAGCGCCGCGTGT......................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ATGGCCGGATGAGGAGA.............................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CGGGTGAGCGCCGCGTTG......................................................................................................................................................................................... | 18 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................GGAGGCGGCCCGCGGCAAT.................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................AGAGCTTGGAGGCGGCCCGCGGGCGAG................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................CCGGATGAGGAGAACC........................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...ATCTGGAAGATCAAGAAGCTCATT............................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CATTAAGAGCTTGGAGGCG................................................................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTGGAAGATCAAGAAGCTCATT............................................................................................................................................................................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCTGGAAGATCAAGAAGCTC.................................................................................................................................................................................................................................. | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GGAAGATCAAGAAGCTCAT................................................................................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ......TGGAAGATCAAGAAGCTCATT............................................................................................................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AGAAGCTCATTAAGAGC......................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GAAGATCAAGAAGCTC.................................................................................................................................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................CAGGGGCGAGGGTCG......................................... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| .....CTGGAAGATCAAGAAG..................................................................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ..................AAGCTCATTAAGAGC......................................................................................................................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAGATCTGGAAGATCAAGAAGCTCATTAAGAGCTTGGAGGCGGCCCGCGGGTGAGCGCCGCGTCCGGCACCCTCCGTGCCCTCTTGGTTCCCGGACCCCTCAGGATGGACGCCCCCGCGCCGCATGGCCGGATGAGGAGAACCAGGCACTAGCCAACTCCCGCCTTCTGGATCTCTGGGTAGTAGCCCCTGACCCAGGGGCGAGGGTCGGCGCGGTTGGCCCGAGAAGGGGTCCTTTCCCCGCATGGGGG ..................................................(((.((((.(((((((((((.....))))).((((((((....))))....)))).))))))....)))))))..(((.(((...((....)).(((....)))...))).))).(((((....)))))....................................................................... ..................................................51...................................................................................................................................184................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040042(GSM532927) G428N. (cervix) | SRR553576(SRX182782) source: Testis. (testes) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR444042(SRX128890) Sample 3cDNABarcode: AF-PP-335: ACG CTC TTC C. (skin) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR040043(GSM532928) G428T. (cervix) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR040017(GSM532902) G645T. (cervix) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM956925Ago2(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR444046(SRX128894) Sample 7cDNABarcode: AF-PP-342: ACG CTC TTC C. (skin) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR444047(SRX128895) Sample 27_1cDNABarcode: AF-PP-343: ACG CTC TT. (skin) | SRR444051(SRX128899) Sample 11cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................GTCCGGGAACCAAGAGGGCACGG.......................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AAGGCGGGAGTTGGCTAGTG.................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ACCCAGAGATCCAGAAGGCGGGAGTTGGCTA...................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGAGAACCAGGCACTGGC................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ACGCCCCCGCGCCGCTGTG........................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................CTAGTGCCTGGTTCTC.................................................................................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |