| (8) B-CELL | (3) BRAIN | (4) BREAST | (11) CELL-LINE | (5) CERVIX | (1) FIBROBLAST | (3) LIVER | (2) OTHER | (2) SKIN | (4) UTERUS |
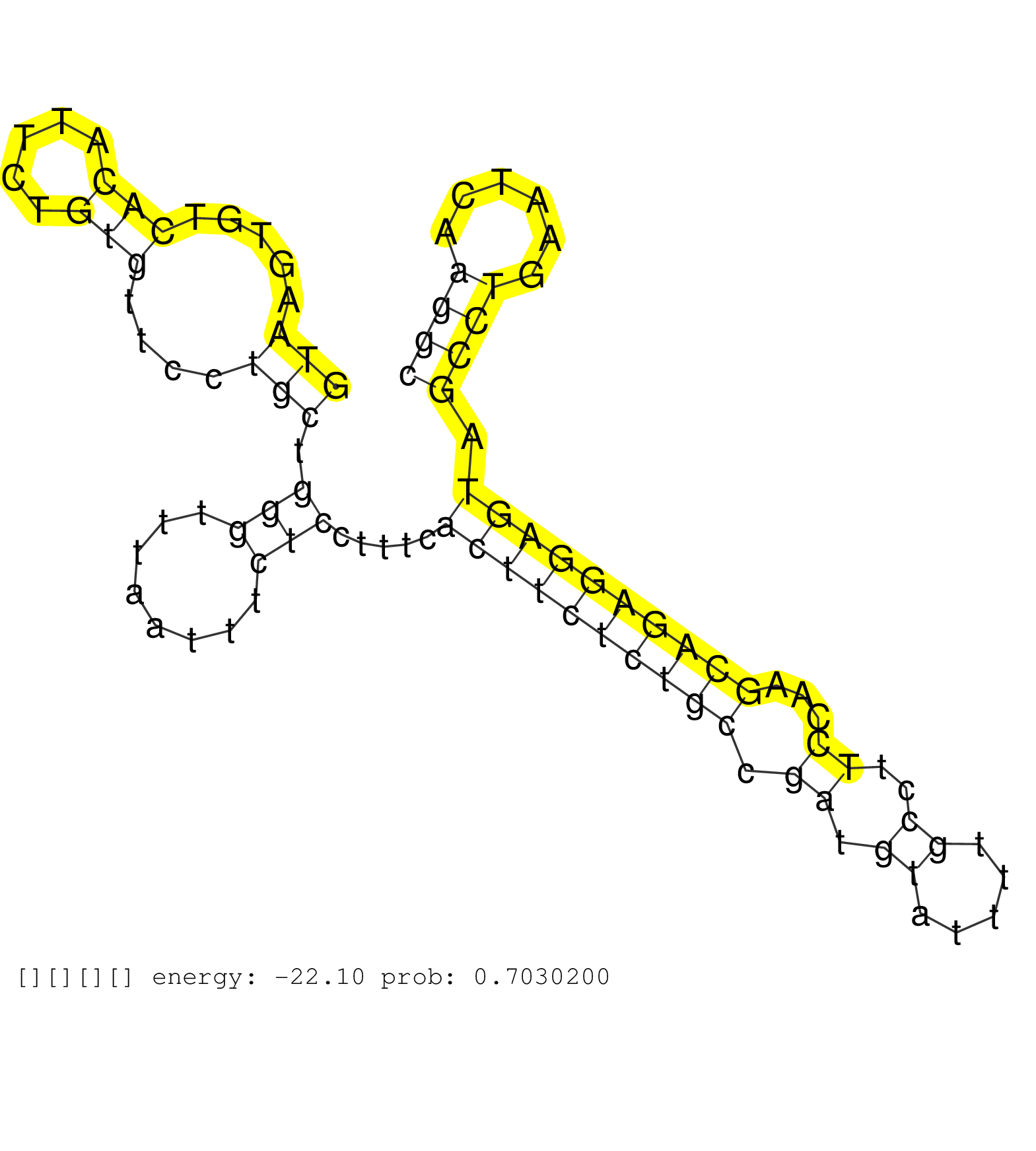
| CACCACTGCACTCCAGCCTGGGCAACAGAGTGAGATACTGTTTCAAAAAAAAAAAAAATCTCATTTACGTTTTTTCTGAAGCGCGTCCTCTTGTGTTACATGGCAAGTCAGGCATCGGCACCGTGCGTATTTTGCAGCGTATATGCTCTACGTGGGGAGCTTTGTAAAAGTGTTTTCTGTTCTGGTACTTCTGTTAACAGCATCACAGCCACAGCTGCCAGTATTGGGGCAGCTGGAATTCCTCAGGCGG .............................................................................(((((((.....(((((........)))))....(((..(((.((.((((((....(((.......)))..)))))).)).))).)))....))))))).....(((..((....))...))).................................................. ........................................................................73.............................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189786 | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189787 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040019(GSM532904) G701T. (cervix) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR040036(GSM532921) G243N. (cervix) | SRR038855(GSM458538) D10. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037938(GSM510476) 293Red. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR040006(GSM532891) G601N. (cervix) | TAX577579(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577738(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR343334 | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................GAAGCGCGTCCTCTTATTT.......................................................................................................................................................... | 19 | 44.00 | 0.00 | 8.00 | - | - | 5.00 | 4.00 | - | 3.00 | 3.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTCTTT.......................................................................................................................................................... | 19 | 8.00 | 0.00 | 5.00 | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTATTC.......................................................................................................................................................... | 19 | 3.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTATT........................................................................................................................................................... | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTTTTT.......................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................TGGGGCAGCTGGAATTCCTC...... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GTTTTCTGTTCTGGTACTTCTGT........................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GAAGCGCGTCCTCTTAT............................................................................................................................................................ | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................TTTTCTGAAGCGCGTCCTCTTGTG........................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTCTGAAGCGCGTCCTCTTG............................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGTATTGGGGCAGCTGGAATTCC........ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGCATCACAGCCACAGCTGCCAGTATTGGGGC.................... | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGGGGCAGCTGGAATTCCTCAGG... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................ATCTCATTTACGTTTTTTTT............................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ............................................................................................................................................................................................................................GTATTGGGGCAGCTGGAATTCCTCAGGCGG | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GGGGCAGCTGGAATTCCTCAGGC.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTGGGGCAGCTGGAATTCCTCAGGCGG | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTGGGGCAGCTGGAATTCCTCAGGC.. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGTATTGGGGCAGCTGGAATTC......... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TATTGGGGCAGCTGGAATTCCTCAGGCGG | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGGGGCAGCTGGAATTCC........ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTGGGGCAGCTGGAATTCCTCAGGCG. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGGGGCAGCTGGAATTCCT....... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GAAGCGCGTCCTCTTATCT.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................GGGGAGCTTTGTAAAACT............................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTCTTG.......................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTACTC.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................GGCAGCTGGAATTCCTCAGGCGG | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................GAAGCGCGTCCTCTTATTG.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GAAGCGCGTCCTCTTACTT.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................CTACGTGGGGAGCTTTGTAAAAGTGTTTT.......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GTATTGGGGCAGCTGGAATTCCTCAGG... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GAAGCGCGTCCTCTTGTTT.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .............................................................................GAAGCGCGTCCTCTTATTA.......................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................TGGGCAACAGAGTGAGATAGAC.................................................................................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................GTATTGGGGCAGCTGG.............. | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGGGGCAGCTGGAATT.......... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACCACTGCACTCCAGCCTGGGCAACAGAGTGAGATACTGTTTCAAAAAAAAAAAAAATCTCATTTACGTTTTTTCTGAAGCGCGTCCTCTTGTGTTACATGGCAAGTCAGGCATCGGCACCGTGCGTATTTTGCAGCGTATATGCTCTACGTGGGGAGCTTTGTAAAAGTGTTTTCTGTTCTGGTACTTCTGTTAACAGCATCACAGCCACAGCTGCCAGTATTGGGGCAGCTGGAATTCCTCAGGCGG .............................................................................(((((((.....(((((........)))))....(((..(((.((.((((((....(((.......)))..)))))).)).))).)))....))))))).....(((..((....))...))).................................................. ........................................................................73.............................................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189786 | SRR189784 | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR189787 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577744(Rovira) total RNA. (breast) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR040019(GSM532904) G701T. (cervix) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | GSM532874(GSM532874) G699T. (cervix) | SRR040036(GSM532921) G243N. (cervix) | SRR038855(GSM458538) D10. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037938(GSM510476) 293Red. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR040006(GSM532891) G601N. (cervix) | TAX577579(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577738(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR343334 | SRR343335 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........CTCCAGCCTGGGCAACAGAGTGAGATCGCC.................................................................................................................................................................................................................. | 30 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............CAGCCTGGGCAACAGAGTGAGATAAAA.................................................................................................................................................................................................................. | 27 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................GTTTCAAAAAAAAAAAAAATTTGC........................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................GTTTCAAAAAAAAAAAAAATCGGGC.......................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................GCAAGTCAGGCATCGGCTT................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .............................................AAAAAAAAAAAAATCTCATCGAG...................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............CCAGCCTGGGCAACAGAGTGAGATGCC................................................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................CTGGGCAACAGAGTGAGATCCCA.................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................AAAAAAAAAAAATCTCATGGAG...................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................CCTGGGCAACAGAGTGAGATATC................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................CCACAGCTGCCAGTATG......................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................CTGGGCAACAGAGTGAGATAC.................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................TTCAAAAAAAAAAAAAATCTGG........................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................TCAAAAAAAAAAAAAATCTCATTGAGG..................................................................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................AAAAAAAAAAAATCTCATCTGG...................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............GCCTGGGCAACAGAGTGAGATCAA................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............GCCTGGGCAACAGAGTGAGATCAAA.................................................................................................................................................................................................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................TTCAAAAAAAAAAAAAATGAGG........................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...............................................................................................................................................................GAAAACACTTTTACAAAG......................................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ...............................................................................................................................................................AAAACACTTTTACAAAG.......................................................................... | 17 | 9 | 0.22 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 |