| (1) B-CELL | (2) BREAST | (7) CELL-LINE | (2) CERVIX | (1) HEART | (2) LIVER | (1) OTHER | (3) SKIN | (1) TESTES |
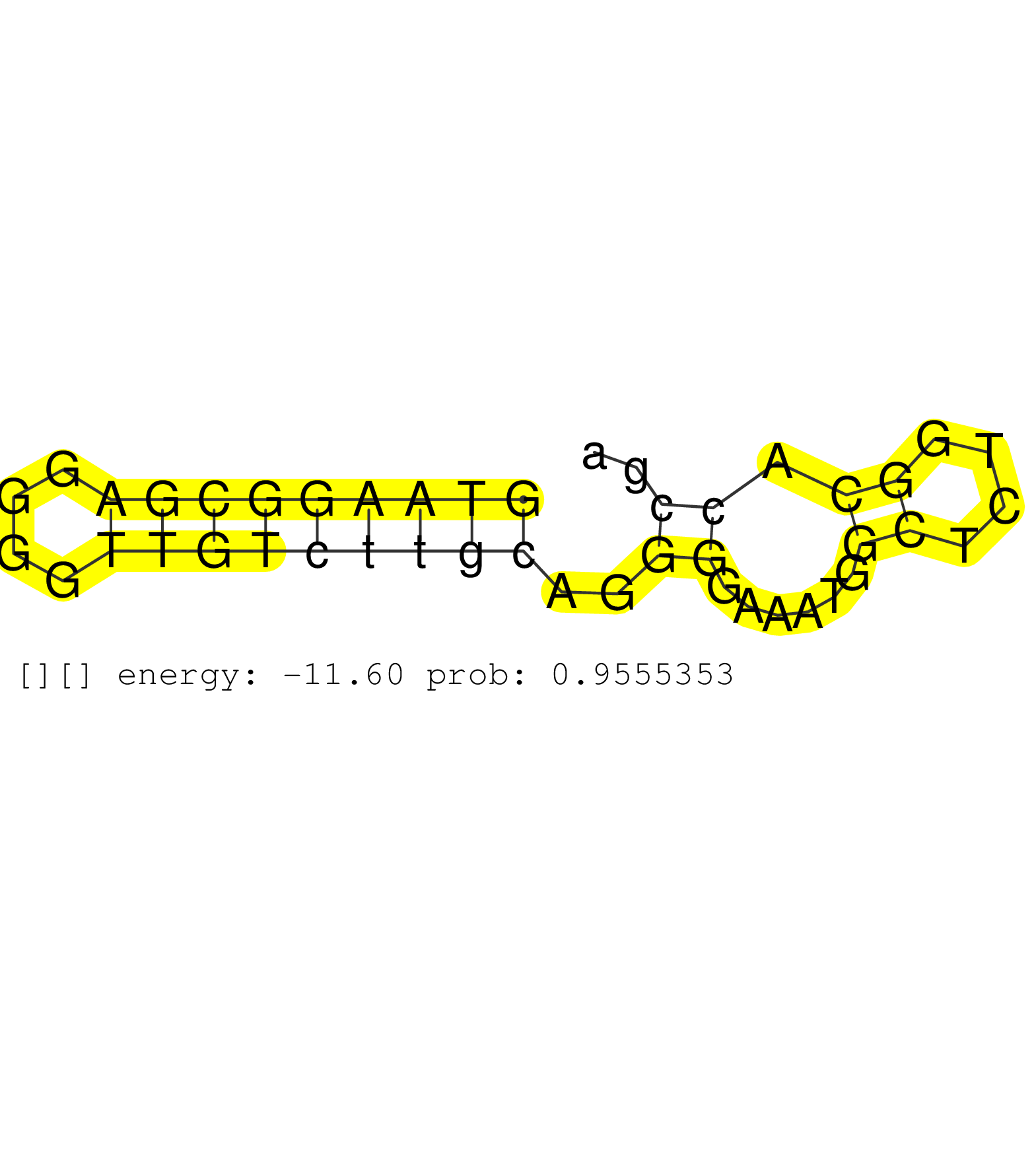
| GCTGTTTTTAAAGGAAAATGTTTTACTTCTGATGGTTTTTATAATTGACAGGTGTATTTGTATGATCTTTGTGTTTGAAGAAAAGAATTGTGCTTAATGTCTGTTTTCTGAATTATAAAGGAAATGACTACATTGTACTTTTTCAGAACGATAAAGCTTTTCATCTCATCTGAGGTCACCTTTTTTTTTTTAATTTTCAGATTTGGGATGATGAAATTGTTGCAGACTTTTACGTAGCTGTTCCAGAAAT ...........................................................................................((((..((((.((..((((((....((((..((((....))))...)))))))))))))))))))).....(((((....))))).......................................................................... .......................................................................................88..............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189782 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR040035(GSM532920) G001T. (cervix) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTAAG................................................... | 21 | 44.00 | 0.00 | 13.00 | 13.00 | 8.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTACTG.................................................. | 22 | 4.00 | 0.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TCACCTTTTTTTTTTTGGG........................................................ | 19 | 3.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTACTT.................................................. | 22 | 3.00 | 0.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTTTTAG................................................. | 23 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTATTG.................................................. | 22 | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTTTTTTTTTTTAATTTTCAG.................................................. | 21 | 3 | 1.67 | 1.67 | 0.33 | 0.67 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CACCTTTTTTTTTTTAATTA...................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTTCTTTG................................................ | 24 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............GAAAATGTTTTACTTCTGGGT........................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..........................................................................................................................................................................................................TTGGGATGATGAAATTGTTGCAGAC....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................GATTTGGGATGATGAAATTGTTGCAGACTTT.................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................GTCACCTTTTTTTTTTTAATTTCCAG.................................................. | 26 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTACCG.................................................. | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTTCAAT................................................. | 23 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TCACCTTTTTTTTTTTAATTTTATTT................................................. | 26 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTTTTTTTTTTTAATTTTCAACA................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGATTTGGGATGATGAAATTGTTGCAGA........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTTCTT.................................................. | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TCACCTTTTTTTTTTTAGCTG...................................................... | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CCTTTTTTTTTTTAATTTATG................................................... | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................ATGAAATTGTTGCAGAC....................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................ATGAAATTGTTGCAGACTT..................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CTTAATGTCTGTTTTTGTC........................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................CACCTTTTTTTTTTTAATCCAG.................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................GTCACCTTTTTTTTTTTACC........................................................ | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................GGGATGATGAAATTGTTGCA.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................ATGAAATTGTTGCAGACTTTTACGTAGCT........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................CTTTTTTTTTTTAATTTTCCGG................................................. | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CTTTTTTTTTTTAATTTTCCCAG................................................ | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................ATAAAGCTTTTCATCT.................................................................................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| GCTGTTTTTAAAGGAAAATGTTTTACTTCTGATGGTTTTTATAATTGACAGGTGTATTTGTATGATCTTTGTGTTTGAAGAAAAGAATTGTGCTTAATGTCTGTTTTCTGAATTATAAAGGAAATGACTACATTGTACTTTTTCAGAACGATAAAGCTTTTCATCTCATCTGAGGTCACCTTTTTTTTTTTAATTTTCAGATTTGGGATGATGAAATTGTTGCAGACTTTTACGTAGCTGTTCCAGAAAT ...........................................................................................((((..((((.((..((((((....((((..((((....))))...)))))))))))))))))))).....(((((....))))).......................................................................... .......................................................................................88..............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189776(GSM714636) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189775(GSM714635) cell line: HEK293clip variant: CLIPenzymatic . (cell line) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189782 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191398(GSM715508) 35genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR040035(GSM532920) G001T. (cervix) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................CATCTCATCTGAGGTCGGGT..................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................TTTGAAGAAAAGAATTAG............................................................................................................................................................... | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................ATGATGAAATTGTTGAAT......................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................TACATTGTACTTTTTCACAG...................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................TCTCATCTGAGGTCACATTG................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| .................................................................................................................................................................CATCTCATCTGAGGTCACGGGC................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................ACATTGTACTTTTTCATTAG..................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................TACATTGTACTTTTTCACTG...................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................TACATTGTACTTTTTCACAC...................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................ACATTGTACTTTTTCATTAC..................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |