| (1) B-CELL | (1) BRAIN | (17) BREAST | (12) CELL-LINE | (7) CERVIX | (1) FIBROBLAST | (3) HEART | (1) KIDNEY | (8) LIVER | (1) OTHER | (1) RRP40.ip | (6) SKIN | (1) TESTES | (4) UTERUS |
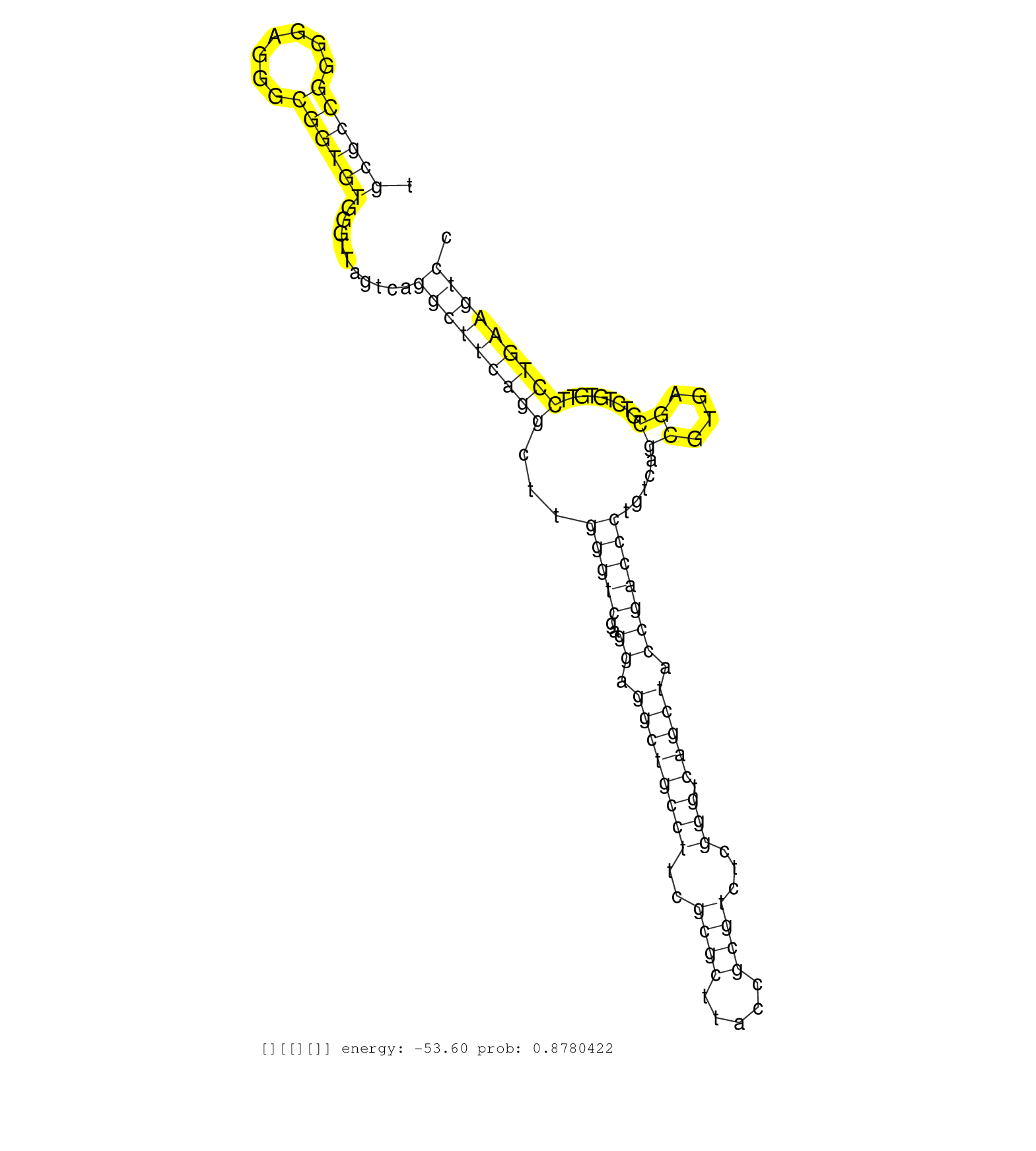
| CAGAAGATACAGATCTGGCAGAAAACTGTGAAGGAGGCTCATTTGACAAAGTGAGTATTTTTTACAAGTAACAGTTTGTGTTTTTTGTGAAACTCCATGGAGAAACAAATTAGTCTGCACATTTTTACAGCTGTGCTAAAGTAATAAAAAATGTAAGGCTGGGCGCGGTGGCTCACGCCTATAATCCCAGCACTTTGGGAGGCCGAGGTGGGTGGATCACCTGAGGTCAGGAGTTTGAGACCAGCCTAGC ...............................................................................................................(((((..((((((((...(((.......)))....)))))))).)))))(((((.((....)).)))))....((((((....)))))).................................................. .......................................................................................................104.............................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577740(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577590(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | TAX577739(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR040042(GSM532927) G428N. (cervix) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040010(GSM532895) G529N. (cervix) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM532929(GSM532929) G702N. (cervix) | GSM532871(GSM532871) G652N. (cervix) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM532874(GSM532874) G699T. (cervix) | SRR189782 | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040008(GSM532893) G727N. (cervix) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR029126(GSM416755) 143B. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR191619(GSM715729) 166genomic small RNA (size selected RNA from . (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR191582(GSM715692) 95genomic small RNA (size selected RNA from t. (breast) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | TAX577743(Rovira) total RNA. (breast) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | GSM532890(GSM532890) G576T. (cervix) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................ATTAGTCTGCACATTGGTT........................................................................................................................... | 19 | 49.00 | 0.00 | - | 5.00 | 5.00 | 8.00 | 5.00 | 3.00 | - | 1.00 | 3.00 | - | - | - | 2.00 | 2.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGGTT........................................................................................................................... | 21 | 18.00 | 0.00 | - | 1.00 | 3.00 | - | 1.00 | 1.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTTGTT........................................................................................................................... | 21 | 3.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGGT............................................................................................................................ | 18 | 3.00 | 0.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................AGGTCAGGAGTTTGAGACCAGCCGCCC | 27 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTCGTT........................................................................................................................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGGCT........................................................................................................................... | 19 | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..GAAGATACAGATCTGGCAGAA................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................AGGTCAGGAGTTTGAGACCAGCCGTCC | 27 | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................TAGTCTGCACATTTTTTA.......................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGGGT........................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................GCGCGGTGGCTCACGCCTATAATCCCAGCCT......................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................TAGTCTGCACATTTTTTAA......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................TCACGCCTATAATCCCAGCACTAAA..................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................AGTCTGCACATTTTTTAA......................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGGT............................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................CCTGAGGTCAGGAGTTTGAGACACGA..... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................AGGAGGCTCATTTGACAA......................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCAGGAGTTTGAGACCAGCCGCCC | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................AGGTCAGGAGTTTGAGACCAGCCGGTC | 27 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGGAT........................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................GAGGTCAGGAGTTTGAGACCAGCCGGTC | 28 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGTTT........................................................................................................................... | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............ATCTGGCAGAAAACTGTGAAGGAGGCTCACTTG............................................................................................................................................................................................................. | 33 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................AAAACTGTGAAGGAGGCTCATCTG............................................................................................................................................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGG............................................................................................................................. | 17 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................AGAAAACTGTGAAGGAGGCT................................................................................................................................................................................................................... | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AATTAGTCTGCACATTGGT............................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGGTG........................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................TAGTCTGCACATTTTTTAT......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............TCTGGCAGAAAACTGTGAAGGAA...................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................AATTAGTCTGCACATTGGTA........................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTTGCC........................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................CAAATTAGTCTGCACATTGTTT........................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................ACCTGAGGTCAGGAGTTTGAGACCAGCCGGTC | 32 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................AATTAGTCTGCACATTGTTT........................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................AGGAGGCTCATTTGACAAAG....................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................CCAGCACTTTGGGAGGCCGAGGTGGGTTGAA................................. | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGACT........................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................TGAGGTCAGGAGTTTGAGACCAGCCGTCC | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGCTT........................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TTACAGCTGTGCTAAAGTAATAAAAAAT.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GTGGGTGGATCACCTGAGGTCAGGAGTTTAAG........... | 32 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................ATTAGTCTGCACATTGTTT........................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................TCAGGAGTTTGAGACCAGCCTATTAT | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................AAATTAGTCTGCACATTGGCT........................................................................................................................... | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................AATTAGTCTGCACATTGCTT........................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................GAAAACTGTGAAGGAGGCT................................................................................................................................................................................................................... | 19 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ..............................................................................................................TAGTCTGCACATTTTTA........................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ................GGCAGAAAACTGTGAAGGAGGCTCATT............................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ..........AGATCTGGCAGAAAACTGTGAAGGAGG..................................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| .......................AACTGTGAAGGAGGCTCATTT.............................................................................................................................................................................................................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .....................AAAACTGTGAAGGAGGCTC.................................................................................................................................................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................AAAACTGTGAAGGAGGCTCATTTGAC........................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ...........GATCTGGCAGAAAACTGTGAAGGAGGCTC.................................................................................................................................................................................................................. | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| CAGAAGATACAGATCTGGCAGAAAACTGTGAAGGAGGCTCATTTGACAAAGTGAGTATTTTTTACAAGTAACAGTTTGTGTTTTTTGTGAAACTCCATGGAGAAACAAATTAGTCTGCACATTTTTACAGCTGTGCTAAAGTAATAAAAAATGTAAGGCTGGGCGCGGTGGCTCACGCCTATAATCCCAGCACTTTGGGAGGCCGAGGTGGGTGGATCACCTGAGGTCAGGAGTTTGAGACCAGCCTAGC ...............................................................................................................(((((..((((((((...(((.......)))....)))))))).)))))(((((.((....)).)))))....((((((....)))))).................................................. .......................................................................................................104.............................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577740(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577590(Rovira) total RNA. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | TAX577746(Rovira) total RNA. (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | TAX577739(Rovira) total RNA. (breast) | TAX577741(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR040042(GSM532927) G428N. (cervix) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR040010(GSM532895) G529N. (cervix) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | GSM532929(GSM532929) G702N. (cervix) | GSM532871(GSM532871) G652N. (cervix) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM532874(GSM532874) G699T. (cervix) | SRR189782 | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR040008(GSM532893) G727N. (cervix) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR029126(GSM416755) 143B. (cell line) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR191619(GSM715729) 166genomic small RNA (size selected RNA from . (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR191582(GSM715692) 95genomic small RNA (size selected RNA from t. (breast) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | TAX577453(Rovira) total RNA. (breast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR553574(SRX182780) source: Heart. (Heart) | TAX577743(Rovira) total RNA. (breast) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | GSM532890(GSM532890) G576T. (cervix) | SRR553576(SRX182782) source: Testis. (testes) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TGGGCGCGGTGGCTCACGCCTATAATCC............................................................... | 28 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................TGGGCGCGGTGGCTCACGCCTATCC.................................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................GGTGGGTGGATCACCTGAGGTCAGGAGTTTGG............ | 32 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................GGAGGCCGAGGTGGGTGGATCACCTGAGGTAG..................... | 32 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................GGCTGGGCGCGGTGGCTCACGCCTAGGAC................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AAGTAATAAAAAATGCAAG............................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................GGCTGGGCGCGGTGGCTCACGCCTATGTG................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................AGCACTTTGGGAGGCCGAGGTGGGTTTTC................................. | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................TGGGCGCGGTGGCTCACGCCTATCCC................................................................. | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |